Spliceosomopathies: Diseases and mechanisms
- PMID: 32506634
- PMCID: PMC8603363
- DOI: 10.1002/dvdy.214
Spliceosomopathies: Diseases and mechanisms
Abstract
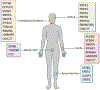
The spliceosome is a complex of RNA and proteins that function together to identify intron-exon junctions in precursor messenger-RNAs, splice out the introns, and join the flanking exons. Mutations in any one of the genes encoding the proteins that make up the spliceosome may result in diseases known as spliceosomopathies. While the spliceosome is active in all cell types, with the majority of the proteins presumably expressed ubiquitously, spliceosomopathies tend to be tissue-specific as a result of germ line or somatic mutations, with phenotypes affecting primarily the retina in retinitis pigmentosa, hematopoietic lineages in myelodysplastic syndromes, or the craniofacial skeleton in mandibulofacial dysostosis. Here we describe the major spliceosomopathies, review the proposed mechanisms underlying retinitis pigmentosa and myelodysplastic syndromes, and discuss how this knowledge may inform our understanding of craniofacial spliceosomopathies.
Keywords: mandibulofacial dysostosis; myelodysplastic syndromes; retinitis pigmentosa; spliceosome.
© 2020 Wiley Periodicals LLC.
Figures
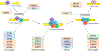
References
-
- Fahim AT, Daiger SP, Weleber RG. Nonsyndromic retinitis pigmentosa overview. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. Gene Reviews. Seattle, WA: University of Washington, Seattle; 1993.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

