Spiking Pandemic Potential: Structural and Immunological Aspects of SARS-CoV-2
- PMID: 32507543
- PMCID: PMC7237910
- DOI: 10.1016/j.tim.2020.05.012
Spiking Pandemic Potential: Structural and Immunological Aspects of SARS-CoV-2
Abstract
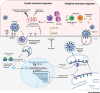
SARS-Coronavirus-2 (SARS-CoV-2) causes Coronavirus disease 2019 (COVID-19), an infectious respiratory disease causing thousands of deaths and overwhelming public health systems. The international spread of SARS-CoV-2 is associated with the ease of global travel, and societal dynamics, immunologic naiveté of the host population, and muted innate immune responses. Based on these factors and the expanding geographic scale of the disease, the World Health Organization (WHO) declared the COVID-19 outbreak a pandemic-the first caused by a coronavirus. In this review, we summarize the current epidemiological status of COVID-19 and consider the virological and immunological lessons, animal models, and tools developed in response to prior SARS-CoV and MERS-CoV outbreaks that can serve as resources for development of SARS-CoV-2 therapeutics and vaccines. In particular, we discuss structural insights into the SARS-CoV-2 spike protein, a major determinant of transmissibility, and discuss key molecular aspects that will aid in understanding and fighting this new global threat.
Keywords: COVID-19; SARS-CoV-2; coronavirus; host immune response; pandemic; spike.
Copyright © 2020 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

