Classification of gastric cancer by EBV status combined with molecular profiling predicts patient prognosis
- PMID: 32508039
- PMCID: PMC7240851
- DOI: 10.1002/ctm2.32
Classification of gastric cancer by EBV status combined with molecular profiling predicts patient prognosis
Abstract
Purpose: To identify how Epstein-Barr virus (EBV) status combined with molecular profiling predicts the prognosis of gastric cancer patients and their associated clinical actionable biomarkers.
Experimental design: A next-generation sequencing assay targeting 295 cancer-related genes was performed in 73 EBV-associated gastric cancer (EBVaGC) and 75 EBV-negative gastric cancer (EBVnGC) specimens and these results were compared with overall survival (OS).
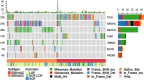
Results: PIK3CA, ARID1A, SMAD4, and PIK3R1 mutated significantly more frequently in EBVaGC compared with their corresponding mutation rate in EBVnGC. As the most frequently mutated gene in EBVnGC (62.7%), TP53 also displayed a mutation rate of 15.1% in EBVaGC. PIK3R1 was revealed as a novel mutated gene (11.0%) associated almost exclusively with EBVaGC. PIK3CA, SMAD4, PIK3R1, and BCOR were revealed to be unique driver genes in EBVaGC. ARID1A displayed a significantly large proportion of inactivated variants in EBVaGC. A notable finding was that integrating the EBV status with tumor mutation burden (TMB) and large genomic instability (LGI) categorized the tumors into four distinct molecular subtypes and optimally predicted patient prognosis. The corresponding median OSs for the EBV+/TMB-high, EBV+/TMB-low, EBV-/LGI-, and EBV-/LGI+ subtypes were 96.2, 75.3, 44.4, and 20.2 months, respectively. The different subtypes were significantly segregated according to distinct mutational profiles and pathways.
Conclusions: Novel mutations in PIK3R1 and TP53 genes, driver genes such as PIK3CA, SMAD4, PIK3R1, BCOR, and ARID1A, and distinguished genomic profiles from EBVnGC were identified in EBVaGC tumors. The classification of gastric cancer by EBV, TMB, and LGI could be a good prognostic indicator, and provides distinguishing, targetable markers for treatment.
Keywords: EBV-associated gastric cancer; copy number variation; prognosis; tumor mutation burden.
© 2020 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Figures


References
-
- Burke AP, Yen TS, Shekitka KM, et al. Lymphoepithelial carcinoma of the stomach with Epstein‐Barr virus demonstrated by polymerase chain reaction. Mod Pathol. 1990;3:377‐380. - PubMed
-
- Qiu MZ, He CY, Lu SX, et al. Prospective observation: clinical utility of plasma Epstein‐Barr virus DNA load in EBV‐associated gastric carcinoma patients. Int J Cancer. 2020;146:272‐280. - PubMed
Grants and funding
- 81602066/National Natural Science Foundation of China
- 81602426/National Natural Science Foundation of China
- 81772587/National Natural Science Foundation of China
- 81802950/National Natural Science Foundation of China
- the third outstanding young talents training plan and Medical Scientist program of Sun Yat-sen University Cancer Center
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
