Tracing the Origins of the Pituitary Adenylate-Cyclase Activating Polypeptide (PACAP)
- PMID: 32508559
- PMCID: PMC7251081
- DOI: 10.3389/fnins.2020.00366
Tracing the Origins of the Pituitary Adenylate-Cyclase Activating Polypeptide (PACAP)
Erratum in
-
Corrigendum: Tracing the Origins of the Pituitary Adenylate-Cyclase Activating Polypeptide (PACAP).Front Neurosci. 2020 Aug 18;14:801. doi: 10.3389/fnins.2020.00801. eCollection 2020. Front Neurosci. 2020. PMID: 33013283 Free PMC article.
Abstract
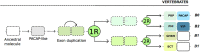
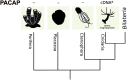
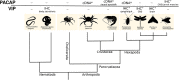
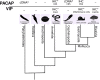
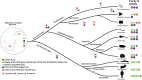
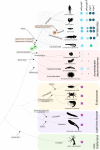
Pituitary adenylate cyclase activating polypeptide (PACAP) is a well-conserved neuropeptide characteristic of vertebrates. This pluripotent hypothalamic neuropeptide regulates neurotransmitter release, intestinal motility, metabolism, cell division/differentiation, and immunity. In vertebrates, PACAP has a specific receptor (PAC1) but it can also activate the Vasoactive Intestinal Peptide receptors (VPAC1 and VPAC2). The evolution of the vertebrate PACAP ligand - receptor pair has been well-described. In contrast, the situation in invertebrates is much less clear. The PACAP ligand - receptor pair in invertebrates has mainly been studied using heterologous antibodies raised against mammalian peptides. A few partial PACAP cDNA clones sharing >87% aa identity with vertebrate PACAP have been isolated from a cnidarian, several protostomes and tunicates but no gene has been reported. Moreover, current evolutionary models of the peptide and receptors using molecular data from phylogenetically distinct invertebrate species (mostly nematodes and arthropods) suggests the PACAP ligand and receptors are exclusive to vertebrate genomes. A basal deuterostome, the cephalochordate amphioxus (Branchiostoma floridae), is the only invertebrate in which elements of a PACAP-like system exists but the peptides and receptor share relatively low sequence conservation with the vertebrate homolog system and are a hybrid with the vertebrate glucagon system. In this study, the evolution of the PACAP system is revisited taking advantage of the burgeoning sequence data (genome and transcriptomes) available for invertebrates to uncover clues about when it first appeared. The results suggest that elements of the PACAP system are absent from protozoans, non-bilaterians, and protostomes and they only emerged after the protostome-deuterostome divergence. PACAP and its receptors appeared in vertebrate genomes and they probably shared a common ancestral origin with the cephalochordate PACAP/GCG-like system which after the genome tetraploidization events that preceded the vertebrate radiation generated the PACAP ligand and receptor pair and also the other members of the Secretin family peptides and their receptors.
Keywords: deuterostomes; early metazoan; evolution; neuropeptide; protostomes; receptor.
Copyright © 2020 Cardoso, Garcia and Power.
Figures




References
-
- Andriès J. C., Belemtougri G., Tramu G. (1991). Multiple peptide immunoreactivities in the nervous system of Aeschna cyanea (Insecta, Odonata) – an immunohistochemical study using antisera to cholecystokinin octapeptide, somatoliberin, vasoactive intestinal peptide, motilin and proctolin. Histochemistry 96 139–148. 10.1007/BF00315984 - DOI - PubMed
LinkOut - more resources
Full Text Sources

