Codon Usage Bias Analysis of Bluetongue Virus Causing Livestock Infection
- PMID: 32508755
- PMCID: PMC7248248
- DOI: 10.3389/fmicb.2020.00655
Codon Usage Bias Analysis of Bluetongue Virus Causing Livestock Infection
Abstract
Bluetongue virus (BTV) is a double-stranded RNA virus with multiple segments and belongs to the genus Orbivirus within the family Reoviridae. BTV is spread to livestock through its dominant vector, biting midges of genus Culicoides. Although great progress has been made in genomic analyses, it is not fully understood how BTVs adapt to their hosts and evade the host's immune systems. In this study, we retrieved BTV genome sequences from the National Center for Biotechnology Information (NCBI) database and performed a comprehensive research to explore the codon usage patterns in 50 BTV strains. We used bioinformatic approaches to calculate the relative synonymous codon usage (RSCU), codon adaptation index (CAI), effective number of codons (ENC), and other indices. The results indicated that most of the overpreferred codons had A-endings, which revealed that mutational pressure was the major force shaping codon usage patterns in BTV. However, the influence of natural selection and geographical factors cannot be ignored on viral codon usage bias. Based on the RSCU values, we performed a comparative analysis between BTVs and their hosts, suggesting that BTVs were inclined to evolve their codon usage patterns that were comparable to those of their hosts. Such findings will be conducive to understanding the elements that contribute to viral evolution and adaptation to hosts.
Keywords: Culicoides; Reoviridae; bluetongue virus; codon usage bias; evolution; nucleotide composition.
Copyright © 2020 Yao, Fan, Yao, Lu, Rahman, Chen and Tao.
Figures

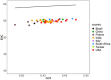
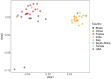
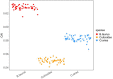

References
LinkOut - more resources
Full Text Sources

