Differential expression profiles of circRNAs in human prostate cancer based on chip and bioinformatic analysis
- PMID: 32509077
- PMCID: PMC7270690
Differential expression profiles of circRNAs in human prostate cancer based on chip and bioinformatic analysis
Abstract
Background: Increasing evidence suggests that circRNAs are involved in the pathogenesis of multiple kinds of cancer. Nevertheless, the differential expression of circRNAs in prostate cancer (PCA) is rarely reported.
Material/method: In our present analyses, circRNAs expression profiles were identified in PCA, based on 5 pairs of PCA and matched non-PCA tissues using circRNA chips.
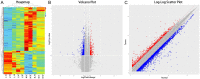
Results: A number of 749 differential circRNAs were expressed between PCA tumor and paracancerous tissues (Fold Change, FC ≥ 2.0 and P < 0.05): 261 were upregulated, whereas 487 were downregulated in PCA tissues. Gene ontology and KEGG pathway analyses indicated that many of the circRNAs are related to carcinogenesis. Circ_0033074 and circ_0016064 both showed changes of maximum magnitude among differentially expressed circRNAs.
Conclusions: Our study detected a relative comprehensive differential map of circRNAs in PCA, which may become novel biomarkers for diagnosis, treatment and follow-up in the future.
Keywords: Prostate cancer; circRNA; expression; microarray.
IJCEP Copyright © 2020.
Conflict of interest statement
None.
Figures

References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. - PubMed
-
- Zheng RS, Sun KX, Zhang SW, Zeng HM, Zou XN, Chen R, Gu XY, Wei WW, He J. Report of cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi. 2019;41:19–28. - PubMed
-
- Tilki D, Schaeffer EM, Evans CP. Understanding mechanisms of resistance in metastatic castration-resistant prostate cancer: the role of the androgen receptor. Eur Urol Focus. 2016;2:499–505. - PubMed
LinkOut - more resources
Full Text Sources
