Emerging roles of ATG proteins and membrane lipids in autophagosome formation
- PMID: 32509328
- PMCID: PMC7248066
- DOI: 10.1038/s41421-020-0161-3
Emerging roles of ATG proteins and membrane lipids in autophagosome formation
Abstract
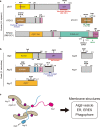
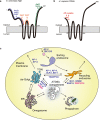
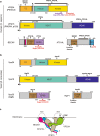
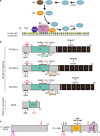
Autophagosome biogenesis is a dynamic membrane event, which is executed by the sequential function of autophagy-related (ATG) proteins. Upon autophagy induction, a cup-shaped membrane structure appears in the cytoplasm, then elongates sequestering cytoplasmic materials, and finally forms a closed double membrane autophagosome. However, how this complex vesicle formation event is strictly controlled and achieved is still enigmatic. Recently, there is accumulating evidence showing that some ATG proteins have the ability to directly interact with membranes, transfer lipids between membranes and regulate lipid metabolism. A novel role for various membrane lipids in autophagosome formation is also emerging. Here, we highlight past and recent key findings on the function of ATG proteins related to autophagosome biogenesis and consider how ATG proteins control this dynamic membrane formation event to organize the autophagosome by collaborating with membrane lipids.
Keywords: Endoplasmic reticulum; Macroautophagy.
© The Author(s) 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

