Epigenetic diversity of Saccharum spp. accessions assessed by methylation-sensitive amplification polymorphism (MSAP)
- PMID: 32509498
- PMCID: PMC7246279
- DOI: 10.1007/s13205-020-02257-7
Epigenetic diversity of Saccharum spp. accessions assessed by methylation-sensitive amplification polymorphism (MSAP)
Abstract
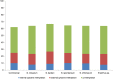
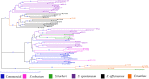
The epigenetic diversity of six genotype groups (commercial cultivars, S. officinarum, S. spontaneum, S. robustum, S. barberi, and Erianthus sp.) was assessed through methylation-sensitive amplification polymorphism (MSAP). A total of 1341 MSAP loci were analyzed, of which 1117 (83.29%) were susceptible to cytosine methylation and responsible for a higher proportion of overall diversity among genotypes. The MSAP selective primer combinations captured different proportions of internal and external cytosine methylation loci across genotype groups, while the average external cytosine frequency was higher for all genotype groups. The genotypes were divided into two subpopulations with a high differentiation index (φst = 0.086) based on epigenetic loci. The genotypes were clustered in three subgroups for both methylated and unmethylated loci, considering dissimilarity values. Four methylated fragments (MFs) were randomly selected and subsequently sequenced and compared with sugarcane public databases using BLASTN. MF alignments suggest that cytosine methylation occurs in sugarcane near CpG islands and tandem repeats within transcribed regions and putative cis-regulatory sequences, which assigned functions are associated with stress adaptation. These results provide the first insights about the distribution of this epigenetic mark in sugarcane genome, and suggest a biological relevance of methylated loci.
Keywords: Cytosine methylation; Epigenome diversity; Saccharum spp.; Wild accessions.
© King Abdulaziz City for Science and Technology 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest in the publication.
Figures


Similar articles
-
Sugarcane mosaic virus mediated changes in cytosine methylation pattern and differentially transcribed fragments in resistance-contrasting sugarcane genotypes.PLoS One. 2020 Nov 9;15(11):e0241493. doi: 10.1371/journal.pone.0241493. eCollection 2020. PLoS One. 2020. PMID: 33166323 Free PMC article.
-
Genetic diversity and population structure analysis of Saccharum and Erianthus genera using microsatellite (SSR) markers.Sci Rep. 2019 Jan 23;9(1):395. doi: 10.1038/s41598-018-36630-7. Sci Rep. 2019. PMID: 30674931 Free PMC article.
-
MSAP markers and global cytosine methylation in plants: a literature survey and comparative analysis for a wild-growing species.Mol Ecol Resour. 2016 Jan;16(1):80-90. doi: 10.1111/1755-0998.12426. Epub 2015 May 18. Mol Ecol Resour. 2016. PMID: 25944158
-
Analysis of sulphur and chlorine induced DNA cytosine methylation alterations in fresh corn (Zea mays L. saccharata and rugosa) leaf tissues by methylation sensitive amplification polymorphism (MSAP) approach.Genes Genomics. 2018 Sep;40(9):913-925. doi: 10.1007/s13258-018-0685-1. Epub 2018 Apr 4. Genes Genomics. 2018. PMID: 30155706
-
Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana.Cytogenet Genome Res. 2005;109(1-3):27-33. doi: 10.1159/000082378. Cytogenet Genome Res. 2005. PMID: 15753555 Review.
Cited by
-
Exploring japonica rice epigenetic diversity in the main production regions of Heilongjiang Province.Sci Rep. 2022 Mar 17;12(1):4592. doi: 10.1038/s41598-022-08683-2. Sci Rep. 2022. PMID: 35301398 Free PMC article.
References
-
- Aitken K, McNeil M. Diversity analysis. In: Henry R, Kole MC, editors. Genetics, genomics and breeding of sugarcane. 1. New York: Science Publishers; 2010. pp. 19–42.
-
- Aitken KS, Li JC, Jackson P, Piperidis G, McIntyre CL. AFLP analysis of genetic diversity within Saccharum officinarum and comparison with sugarcane cultivars. Aust J Agric Res. 2006;57(11):1167–1184. doi: 10.1071/AR05391. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

