Gut Microbiota Dysbiosis Associated With Altered Production of Short Chain Fatty Acids in Children With Neurodevelopmental Disorders
- PMID: 32509596
- PMCID: PMC7248180
- DOI: 10.3389/fcimb.2020.00223
Gut Microbiota Dysbiosis Associated With Altered Production of Short Chain Fatty Acids in Children With Neurodevelopmental Disorders
Abstract
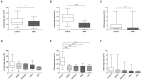
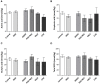
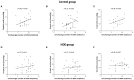
While gut microbiota dysbiosis has been linked with autism, its role in the etiology of other neurodevelopmental disorders (NDD) is largely underexplored. To our knowledge this is the first study to evaluate gut microbiota diversity and composition in 36 children from the Republic of Serbia diagnosed with NDD and 28 healthy children. The results revealed an increased incidence of potentially harmful bacteria, closely related to Clostridium species, in the NDD patient group compared to the Control group: Desulfotomaculum guttoideum (P < 0.01), Intestinibacter bartlettii (P < 0.05), and Romboutsia ilealis (P < 0.001). On the other hand, significantly lower diversity of common commensal bacteria in the NDD group of patients was noticed. Enterococcus faecalis (P < 0.05), Enterococcus gallinarum (P < 0.01), Streptococcus pasteurianus (P < 0.05), Lactobacillus rhamnosus (P < 0.01) and Bifidobacteria sp. were detected in lower numbers of patients or were even absent in some NDD patients. In addition, butyrate-producing bacteria Faecalibacterium prausnitzii (P < 0.01), Butyricicoccus pullicaecorum (P < 0.05), and Eubacterium rectale (P = 0.07) were less frequent in the NDD patient group. In line with that, the levels of fecal short chain fatty acids (SCFAs) were determined. Although significant differences in SCFA levels were not detected between NDD patients and the Control group, a positive correlation was noted between number of rDNA amplicons obtained with universal primers and level of propionic acid, as well as a trend for levels of total SCFAs and butyric acid in the Control group. This correlation is lost in the NDD patient group, indicating that NDD patients' microbiota differs from the microbiota of healthy children in the presence or number of strong SCFA-producing bacteria. According to a range-weighted richness index it was observed that microbial diversity was significantly lower in the NDD patient group. Our study reveals that the intestinal microbiota from NDD patients differs from the microbiota of healthy children. It is hypothesized that early life microbiome might have an impact on GI disturbances and accompanied behavioral problems frequently observed in patients with a broad spectrum of NDD.
Keywords: Bifidobacterium; Clostridium like species; Lactobacillus; autism; gut-brain axis; microbial diversity.
Copyright © 2020 Bojović, Ignjatović, Soković Bajić, Vojnović Milutinović, Tomić, Golić and Tolinački.
Figures


References
-
- Belenguer A., Duncan S. H., Calder A. G., Holtrop G., Louis P., Lobley G. E., et al. (2006). Two routes of metabolic cross-feeding between bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72, 3593–3599. 10.1128/AEM.72.5.3593-3599.2006 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

