ProTrack: An Interactive Multi-Omics Data Browser for Proteogenomic Studies
- PMID: 32510176
- PMCID: PMC7677200
- DOI: 10.1002/pmic.201900359
ProTrack: An Interactive Multi-Omics Data Browser for Proteogenomic Studies
Abstract
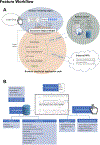
The Clinical Proteomic Tumor Analysis Consortium (CPTAC) initiative has generated extensive multi-omics data resources of deep proteogenomic profiles for multiple cancer types. To enable the broader community of biological and medical researchers to intuitively query, explore, and download data and analysis results from various CPTAC projects, a prototype user-friendly web application called "ProTrack" is built with the CPTAC clear cell renal cell carcinoma (ccRCC) data set (http://ccrcc.cptac-data-view.org). Here the salient features of this application which provides a dynamic, comprehensive, and granular visualization of the rich proteogenomic data is described.
© 2020 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of Interest Statement
The authors have no conflict of interest to declare.
Figures

Comment in
-
A User-Friendly Visualization Tool for Multi-Omics Data.Proteomics. 2020 Nov;20(21-22):e2000136. doi: 10.1002/pmic.202000136. Epub 2020 Sep 3. Proteomics. 2020. PMID: 32744797
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

