Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes
- PMID: 32510327
- PMCID: PMC7311171
- DOI: 10.7554/eLife.55954
Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes
Abstract
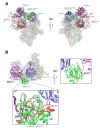
Dynamic trafficking of G protein-coupled receptors (GPCRs) out of cilia is mediated by the BBSome. In concert with its membrane recruitment factor, the small GTPase ARL6/BBS3, the BBSome ferries GPCRs across the transition zone, a diffusion barrier at the base of cilia. Here, we present the near-atomic structures of the BBSome by itself and in complex with ARL6GTP, and we describe the changes in BBSome conformation induced by ARL6GTP binding. Modeling the interactions of the BBSome with membranes and the GPCR Smoothened (SMO) reveals that SMO, and likely also other GPCR cargoes, must release their amphipathic helix 8 from the membrane to be recognized by the BBSome.
Keywords: BBSome; Bardet-Biedl Syndrome; Cilia; GPCR; cell biology; hedgehog signaling; molecular biophysics; mouse; smoothened; structural biology.
© 2020, Yang et al.
Conflict of interest statement
SY, KB, HC, JW, US, TW, MN No competing interests declared
Figures











References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

