Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis
- PMID: 32510973
- PMCID: PMC7381712
- DOI: 10.1152/ajpcell.00224.2020
Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis
Abstract
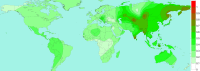
Because of the ongoing pandemic around the world, the mechanisms underlying the SARS-CoV-2-induced COVID-19 are subject to intense investigation. Based on available data for the SARS-CoV-1 virus, we suggest how CoV-2 localization of RNA transcripts in mitochondria hijacks the host cell's mitochondrial function to viral advantage. Besides viral RNA transcripts, RNA also localizes to mitochondria. SARS-CoV-2 may manipulate mitochondrial function indirectly, first by ACE2 regulation of mitochondrial function, and once it enters the host cell, open-reading frames (ORFs) such as ORF-9b can directly manipulate mitochondrial function to evade host cell immunity and facilitate virus replication and COVID-19 disease. Manipulations of host mitochondria by viral ORFs can release mitochondrial DNA (mtDNA) in the cytoplasm and activate mtDNA-induced inflammasome and suppress innate and adaptive immunity. We argue that a decline in ACE2 function in aged individuals, coupled with the age-associated decline in mitochondrial functions resulting in chronic metabolic disorders like diabetes or cancer, may make the host more vulnerable to infection and health complications to mortality. These observations suggest that distinct localization of viral RNA and proteins in mitochondria must play essential roles in SARS-CoV-2 pathogenesis. Understanding the mechanisms underlying virus communication with host mitochondria may provide critical insights into COVID-19 pathologies. An investigation into the SARS-CoV-2 hijacking of mitochondria should lead to novel approaches to prevent and treat COVID-19.
Keywords: COVID-19; SARS-CoV; aging; coronavirus; mitochondria; mitochondrial DNA.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

