CrAssphage as a Novel Tool to Detect Human Fecal Contamination on Environmental Surfaces and Hands
- PMID: 32511090
- PMCID: PMC7392416
- DOI: 10.3201/eid2608.200346
CrAssphage as a Novel Tool to Detect Human Fecal Contamination on Environmental Surfaces and Hands
Abstract
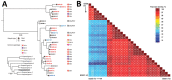
CrAssphage is a recently discovered human gut-associated bacteriophage. To validate the potential use of crAssphage for detecting human fecal contamination on environmental surfaces and hands, we tested stool samples (n = 60), hand samples (n = 30), and environmental swab samples (n = 201) from 17 norovirus outbreaks for crAssphage by real-time PCR. In addition, we tested stool samples from healthy persons (n = 173), respiratory samples (n = 113), and animal fecal specimens (n = 68) and further sequenced positive samples. Overall, we detected crAssphage in 71.4% of outbreak stool samples, 48%-68.5% of stool samples from healthy persons, 56.2% of environmental swabs, and 60% of hand rinse samples, but not in human respiratory samples or animal fecal samples. CrAssphage sequences could be grouped into 2 major genetic clusters. Our data suggest that crAssphage could be used to detect human fecal contamination on environmental surfaces and hands.
Keywords: crAssphage; cruise ship; enteric infections; environmental contamination; hand contamination; human fecal indicator; long-term care facility; norovirus outbreak; viruses.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

