This is a preprint.
Sequence analysis of SARS-CoV-2 genome reveals features important for vaccine design
- PMID: 32511300
- PMCID: PMC7217226
- DOI: 10.1101/2020.03.30.016832
Sequence analysis of SARS-CoV-2 genome reveals features important for vaccine design
Update in
-
Sequence analysis of SARS-CoV-2 genome reveals features important for vaccine design.Sci Rep. 2020 Sep 24;10(1):15643. doi: 10.1038/s41598-020-72533-2. Sci Rep. 2020. PMID: 32973171 Free PMC article.
Abstract
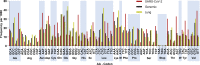
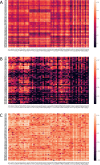
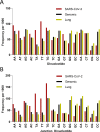
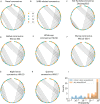
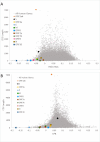
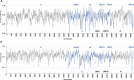
As the SARS-CoV-2 pandemic is rapidly progressing, the need for the development of an effective vaccine is critical. A promising approach for vaccine development is to generate, through codon pair deoptimization, an attenuated virus. This approach carries the advantage that it only requires limited knowledge specific to the virus in question, other than its genome sequence. Therefore, it is well suited for emerging viruses for which we may not have extensive data. We performed comprehensive in silico analyses of several features of SARS-CoV-2 genomic sequence (e.g., codon usage, codon pair usage, dinucleotide/junction dinucleotide usage, RNA structure around the frameshift region) in comparison with other members of the coronaviridae family of viruses, the overall human genome, and the transcriptome of specific human tissues such as lung, which are primarily targeted by the virus. Our analysis identified the spike (S) and nucleocapsid (N) proteins as promising targets for deoptimization and suggests a roadmap for SARS-CoV-2 vaccine development, which can be generalizable to other viruses.
Conflict of interest statement
Conflict of Interest The authors declare no competing interests.
Figures
References
-
- Organization W. H. Coronavirus disease 2019 (COVID-19) Situation Report – 70, <https://www.who.int/docs/default-source/coronaviruse/situation-reports/2...> (2020).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
