This is a preprint.
Analysis of SARS-CoV-2 Antibodies in COVID-19 Convalescent Blood using a Coronavirus Antigen Microarray
- PMID: 32511302
- PMCID: PMC7217240
- DOI: 10.1101/2020.04.15.043364
Analysis of SARS-CoV-2 Antibodies in COVID-19 Convalescent Blood using a Coronavirus Antigen Microarray
Update in
-
Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent blood using a coronavirus antigen microarray.Nat Commun. 2021 Jan 4;12(1):6. doi: 10.1038/s41467-020-20095-2. Nat Commun. 2021. PMID: 33397903 Free PMC article.
Abstract
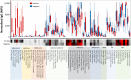
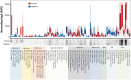
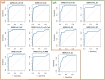
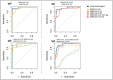
The current practice for diagnosis of COVID-19, based on SARS-CoV-2 PCR testing of pharyngeal or respiratory specimens in a symptomatic patient at high epidemiologic risk, likely underestimates the true prevalence of infection. Serologic methods can more accurately estimate the disease burden by detecting infections missed by the limited testing performed to date. Here, we describe the validation of a coronavirus antigen microarray containing immunologically significant antigens from SARS-CoV-2, in addition to SARS-CoV, MERS-CoV, common human coronavirus strains, and other common respiratory viruses. A comparison of antibody profiles detected on the array from control sera collected prior to the SARS-CoV-2 pandemic versus convalescent blood specimens from virologically confirmed COVID-19 cases demonstrates near complete discrimination of these two groups, with improved performance from use of antigen combinations that include both spike protein and nucleoprotein. This array can be used as a diagnostic tool, as an epidemiologic tool to more accurately estimate the disease burden of COVID-19, and as a research tool to correlate antibody responses with clinical outcomes.
Conflict of interest statement
Competing Interests The coronavirus antigen microarray is intellectual property of the Regents of the University of California that is licensed for commercialization to Nanommune Inc. (Irvine, CA), a private company for which P. Felgner is the largest shareholder and several coauthors (R. de Assis, A. Jain, R. Nakajima, A. Jasinskas, J. Obiero, H. Davies, and S. Khan) also own shares. Nanommune Inc. has a business partnership with Sino Biological Inc. (Beijing, China) which expressed and purified the antigens used in this study. The convalescent plasma used in this study was collected for clinical use by independent blood centers using licensed plasma or platelet processing systems manufactured by Cerus Corporation, for which multiple authors (L. Corash, A. Bagri) are shareholders and employees. Convalescent sera were also provided by Ortho Clinical Diagnostics, which is using these specimens to validate a clinical diagnostic test, and P. Hosimer, C. Noesen, and P. Contestable are shareholders and employees of this company. M. Battegay, A. Buser and A. Holbro are employees of the University of Basel and have no conflicts of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
