This is a preprint.
Coronavirus infection and PARP expression dysregulate the NAD Metabolome: an actionable component of innate immunity
- PMID: 32511303
- PMCID: PMC7217258
- DOI: 10.1101/2020.04.17.047480
Coronavirus infection and PARP expression dysregulate the NAD Metabolome: an actionable component of innate immunity
Update in
-
Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity.J Biol Chem. 2020 Dec 25;295(52):17986-17996. doi: 10.1074/jbc.RA120.015138. Epub 2020 Oct 13. J Biol Chem. 2020. PMID: 33051211 Free PMC article.
Abstract
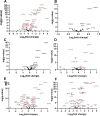
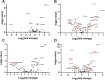
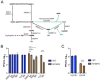
Poly-ADP-ribose polymerase (PARP) superfamily members covalently link either a single ADP-ribose (ADPR) or a chain of ADPR units to proteins using nicotinamide adenine dinucleotide (NAD) as the source of ADPR. While the well-known poly-ADP-ribosylating (PARylating) PARPs primarily function in the DNA damage response, many non-canonical mono-ADP-ribosylating (MARylating) PARPs are associated with cellular antiviral responses. We recently demonstrated robust upregulation of several PARPs following infection with Murine Hepatitis Virus (MHV), a model coronavirus. Here we show that SARS-CoV-2 infection strikingly upregulates MARylating PARPs and induces the expression of genes encoding enzymes for salvage NAD synthesis from nicotinamide (NAM) and nicotinamide riboside (NR), while downregulating other NAD biosynthetic pathways. We show that overexpression of PARP10 is sufficient to depress cellular NAD and that the activities of the transcriptionally induced enzymes PARP7, PARP10, PARP12 and PARP14 are limited by cellular NAD and can be enhanced by pharmacological activation of NAD synthesis. We further demonstrate that infection with MHV induces a severe attack on host cell NAD+ and NADP+. Finally, we show that NAMPT activation, NAM and NR dramatically decrease the replication of an MHV virus that is sensitive to PARP activity. These data suggest that the antiviral activities of noncanonical PARP isozyme activities are limited by the availability of NAD, and that nutritional and pharmacological interventions to enhance NAD levels may boost innate immunity to coronaviruses.
Keywords: ADP-ribosylation; COVID-19; Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); interferon; nicotinamide adenine dinucleotide (NAD); poly(ADP-ribose) polymerase (PARP); transcriptomics.
Conflict of interest statement
COMPETING INTERESTS CB is chief scientific adviser of ChromaDex and owns shares of ChromaDex stock. CB, SAJT, SP and ARF filed an invention disclosure on uses of NAD-boosting with respect to protection against coronavirus infection. Others declare no competing interests.
Figures


References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., China Novel Coronavirus, I., and Research, T. (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733 - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
