This is a preprint.
The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2
- PMID: 32511321
- PMCID: PMC7239051
- DOI: 10.1101/2020.03.16.994236
The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2
Update in
-
Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2.Science. 2020 Sep 4;369(6508):1261-1265. doi: 10.1126/science.abc0870. Epub 2020 Aug 4. Science. 2020. PMID: 32753553 Free PMC article.
Abstract
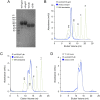
The rapid and escalating spread of SARS coronavirus 2 (SARS-CoV-2) poses an immediate public health emergency. The viral spike protein S binds ACE2 on host cells to initiate molecular events that release the viral genome intracellularly. Soluble ACE2 inhibits entry of both SARS and SARS-2 coronaviruses by acting as a decoy for S binding sites, and is a candidate for therapeutic, prophylactic and diagnostic development. Using deep mutagenesis, variants of ACE2 are identified with increased binding to the receptor binding domain of S. Mutations are found across the interface, in the N90-glycosylation motif, and at buried sites where they are predicted to enhance local folding and presentation of the interaction epitope. When single substitutions are combined, large increases in binding can be achieved. The mutational landscape offers a blueprint for engineering high affinity proteins and peptides that block receptor binding sites on S to meet this unprecedented challenge.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT. E.P. is the inventor on a provisional patent filing by the University of Illinois claiming mutations in ACE2 described here that enhance binding to S. E.P. is a cofounder of Orthogonal Biologics Inc, which has a license from the University of Illinois.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
