This is a preprint.
A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 spike-mediated cell entry and its inhibition
- PMID: 32511365
- PMCID: PMC7263493
- DOI: 10.1101/2020.05.20.105247
A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 spike-mediated cell entry and its inhibition
Update in
-
A Replication-Competent Vesicular Stomatitis Virus for Studies of SARS-CoV-2 Spike-Mediated Cell Entry and Its Inhibition.Cell Host Microbe. 2020 Sep 9;28(3):486-496.e6. doi: 10.1016/j.chom.2020.06.020. Epub 2020 Jul 3. Cell Host Microbe. 2020. PMID: 32738193 Free PMC article.
Abstract
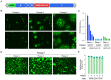
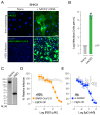
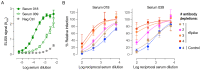
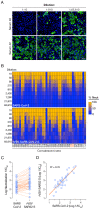
There is an urgent need for vaccines and therapeutics to prevent and treat COVID-19. Rapid SARS-CoV-2 countermeasure development is contingent on the availability of robust, scalable, and readily deployable surrogate viral assays to screen antiviral humoral responses, and define correlates of immune protection, and to down-select candidate antivirals. Here, we describe a highly infectious recombinant vesicular stomatitis virus bearing the SARS-CoV-2 spike glycoprotein S as its sole entry glycoprotein that closely resembles the authentic agent in its entry-related properties. We show that the neutralizing activities of a large panel of COVID-19 convalescent sera can be assessed in high-throughput fluorescent reporter assay with rVSV-SARS-CoV-2 S and that neutralization of the rVSV and authentic SARS-CoV-2 by spike-specific antibodies in these antisera is highly correlated. Our findings underscore the utility of rVSV-SARS-CoV-2 S for the development of spike-specific vaccines and therapeutics and for mechanistic studies of viral entry and its inhibition.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
