This is a preprint.
Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium
- PMID: 32511382
- PMCID: PMC7263511
- DOI: 10.1101/2020.05.06.081695
Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium
Update in
-
Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes.PLoS Biol. 2021 Mar 17;19(3):e3001143. doi: 10.1371/journal.pbio.3001143. eCollection 2021 Mar. PLoS Biol. 2021. PMID: 33730024 Free PMC article.
Abstract
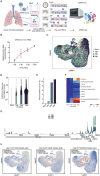
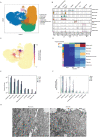
SARS-CoV-2, the causative agent of COVID-19, has tragically burdened individuals and institutions around the world. There are currently no approved drugs or vaccines for the treatment or prevention of COVID-19. Enhanced understanding of SARS-CoV-2 infection and pathogenesis is critical for the development of therapeutics. To reveal insight into viral replication, cell tropism, and host-viral interactions of SARS-CoV-2 we performed single-cell RNA sequencing of experimentally infected human bronchial epithelial cells (HBECs) in air-liquid interface cultures over a time-course. This revealed novel polyadenylated viral transcripts and highlighted ciliated cells as a major target of infection, which we confirmed by electron microscopy. Over the course of infection, cell tropism of SARS-CoV-2 expands to other epithelial cell types including basal and club cells. Infection induces cell-intrinsic expression of type I and type III IFNs and IL6 but not IL1. This results in expression of interferon-stimulated genes in both infected and bystander cells. We observe similar gene expression changes from a COVID-19 patient ex vivo. In addition, we developed a new computational method termed CONditional DENSity Embedding (CONDENSE) to characterize and compare temporal gene dynamics in response to infection, which revealed genes relating to endothelin, angio-genesis, interferon, and inflammation-causing signaling pathways. In this study, we conducted an in-depth analysis of SARS-CoV-2 infection in HBECs and a COVID-19 patient and revealed genes, cell types, and cell state changes associated with infection.
Conflict of interest statement
Declaration of interests: The authors declare no competing interests.
Figures





References
-
- W. H. Organization, Novel Coronavirus(2019-nCoV), Situation Report 22, 2020.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
