This is a preprint.
A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease
- PMID: 32511434
- PMCID: PMC7217088
- DOI: 10.1101/2020.04.14.20065771
A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease
Update in
-
A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity.Nat Commun. 2020 Sep 17;11(1):4704. doi: 10.1038/s41467-020-18450-4. Nat Commun. 2020. PMID: 32943637 Free PMC article.
Abstract
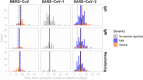
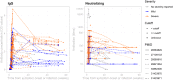
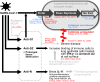
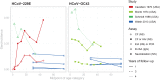
The duration and nature of immunity generated in response to SARS-CoV-2 infection is unknown. Many public health responses and modeled scenarios for COVID-19 outbreaks caused by SARSCoV-2 assume that infection results in an immune response that protects individuals from future infections or illness for some amount of time. The timescale of protection is a critical determinant of the future impact of the pathogen. The presence or absence of protective immunity due to infection or vaccination (when available) will affect future transmission and illness severity. The dynamics of immunity and nature of protection are relevant to discussions surrounding therapeutic use of convalescent sera as well as efforts to identify individuals with protective immunity. Here, we review the scientific literature on antibody immunity to coronaviruses, including SARS-CoV-2 as well as the related SARS-CoV-1, MERS-CoV and human endemic coronaviruses (HCoVs). We reviewed 1281 abstracts and identified 322 manuscripts relevant to 5 areas of focus: 1) antibody kinetics, 2) correlates of protection, 3) immunopathogenesis, 4) antigenic diversity and cross-reactivity, and 5) population seroprevalence. While studies of SARS-CoV-2 are necessary to determine immune responses to it, evidence from other coronaviruses can provide clues and guide future research.
Figures




References
-
- Godlee F. The burning building. BMJ m1101 (2020) doi:10.1136/bmj.m1101 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
