This is a preprint.
Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay
- PMID: 32511449
- PMCID: PMC7239074
- DOI: 10.1101/2020.03.06.20032334
Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay
Abstract
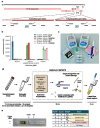
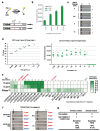
An outbreak of novel betacoronavirus, SARS-CoV-2 (formerly named 2019-nCoV), began in Wuhan, China in December 2019 and the COVID-19 disease associated with infection has since spread rapidly to multiple countries. Here we report the development of SARS-CoV-2 DETECTR, a rapid (~30 min), low-cost, and accurate CRISPR-Cas12 based lateral flow assay for detection of SARS-CoV-2 from respiratory swab RNA extracts. We validated this method using contrived reference samples and clinical samples from infected US patients and demonstrated comparable performance to the US CDC SARS-CoV-2 real-time RT-PCR assay.
Keywords: 2019-nCoV; COVID-19; CRISPR; CRISPR-Cas12; DETECTR; SARS-CoV-2; Wuhan; betacoronavirus; coronavirus; diagnostic testing; epidemic; isothermal amplification; lateral flow; loop-mediated isothermal amplification (LAMP); molecular testing; outbreak; pandemic; zoonotic.
Figures
References
-
- World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report - 44. 8 (World Health Organization, Geneva, Switzerland, 2020).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
