This is a preprint.
Test performance evaluation of SARS-CoV-2 serological assays
- PMID: 32511497
- PMCID: PMC7273265
- DOI: 10.1101/2020.04.25.20074856
Test performance evaluation of SARS-CoV-2 serological assays
Update in
-
Evaluation of SARS-CoV-2 serology assays reveals a range of test performance.Nat Biotechnol. 2020 Oct;38(10):1174-1183. doi: 10.1038/s41587-020-0659-0. Epub 2020 Aug 27. Nat Biotechnol. 2020. PMID: 32855547 Free PMC article.
Abstract
Background: Serological tests are crucial tools for assessments of SARS-CoV-2 exposure, infection and potential immunity. Their appropriate use and interpretation require accurate assay performance data.
Method: We conducted an evaluation of 10 lateral flow assays (LFAs) and two ELISAs to detect anti-SARS-CoV-2 antibodies. The specimen set comprised 128 plasma or serum samples from 79 symptomatic SARS-CoV-2 RT-PCR-positive individuals; 108 pre-COVID-19 negative controls; and 52 recent samples from individuals who underwent respiratory viral testing but were not diagnosed with Coronavirus Disease 2019 (COVID-19). Samples were blinded and LFA results were interpreted by two independent readers, using a standardized intensity scoring system.
Results: Among specimens from SARS-CoV-2 RT-PCR-positive individuals, the percent seropositive increased with time interval, peaking at 81.8-100.0% in samples taken >20 days after symptom onset. Test specificity ranged from 84.3-100.0% in pre-COVID-19 specimens. Specificity was higher when weak LFA bands were considered negative, but this decreased sensitivity. IgM detection was more variable than IgG, and detection was highest when IgM and IgG results were combined. Agreement between ELISAs and LFAs ranged from 75.7-94.8%. No consistent cross-reactivity was observed.
Conclusion: Our evaluation showed heterogeneous assay performance. Reader training is key to reliable LFA performance, and can be tailored for survey goals. Informed use of serology will require evaluations covering the full spectrum of SARS-CoV-2 infections, from asymptomatic and mild infection to severe disease, and later convalescence. Well-designed studies to elucidate the mechanisms and serological correlates of protective immunity will be crucial to guide rational clinical and public health policies.
Conflict of interest statement
Competing Interests This work was supported by gifts from Anthem Blue Cross Blue Shield, the Chan Zuckerberg Biohub, and anonymous philanthropy. C.Y.C. is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center, receives research support funding from Abbott Laboratories and is on the Scientific Advisory Board of Mammoth Biosciences, Inc. C. J. Y. is cofounder of DropPrint Genomics and serves as an advisor to them. M.S.A. holds stock in Medtronic and Merck. P.D.H. is a cofounder of Spotlight Therapeutics and serves on the board of directors and scientific advisory board, and is an advisor to Serotiny. P.D.H. holds stock in Spotlight Therapeutics and Editas Medicine. A.M. is a cofounder of Spotlight Therapeutics and Arsenal Biosciences and serves on their boards of directors and scientific advisory boards. A.M. has served as an advisor to Juno Therapeutics, was a member of the scientific advisory board at PACT Pharma, and was an advisor to Trizell. A.M. owns stock in Arsenal Biosciences, Spotlight Therapeutics and PACT Pharma. RY owns stock in Abbvie, Bluebird Bio, Bristol Myers Squibb, Cara Therapeutics, Editas Medicine, Esperion, and Gilead Sciences. Unrelated to this current work, the Marson lab has received sponsored research support from Juno Therapeutics, Epinomics and Sanofi, and a gift from Gilead.
Figures
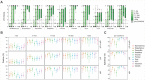
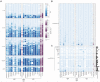
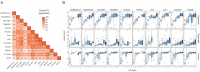
References
-
- Johns Hopkins University. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594.... Published 2020. Accessed April 12, 2020.
-
- Infectious Diseases Society of America. IDSA Statement on COVID-19 Testing. https://www.idsociety.org/globalassets/idsa/public-health/covid-19-idsa-.... Published 2020. Accessed April 20th, 2020.
-
- World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. Geneva: World Health Organization; 2020; 2020.
