This is a preprint.
A Unique Clade of SARS-CoV-2 Viruses is Associated with Lower Viral Loads in Patient Upper Airways
- PMID: 32511558
- PMCID: PMC7274239
- DOI: 10.1101/2020.05.19.20107144
A Unique Clade of SARS-CoV-2 Viruses is Associated with Lower Viral Loads in Patient Upper Airways
Update in
-
A clade of SARS-CoV-2 viruses associated with lower viral loads in patient upper airways.EBioMedicine. 2020 Dec;62:103112. doi: 10.1016/j.ebiom.2020.103112. Epub 2020 Nov 11. EBioMedicine. 2020. PMID: 33186810 Free PMC article. Clinical Trial.
Abstract
Background: The rapid spread of SARS-CoV-2, the causative agent of Coronavirus disease 2019 (COVID-19), has been accompanied by the emergence of distinct viral clades, though their clinical significance remains unclear. Here, we aimed to investigate the phylogenetic characteristics of SARS-CoV-2 infections in Chicago, Illinois and assess their relationship to clinical parameters.
Methods: We performed whole-genome sequencing of SARS-CoV-2 isolates collected from COVID-19 patients in a Chicago healthcare system in mid-March, 2020. Using these and other publicly available sequences, we performed phylogenetic, phylogeographic, and phylodynamic analyses. Patient data was assessed for correlations between demographic or clinical characteristics and virologic features.
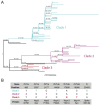
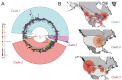
Findings: The 88 SARS-CoV-2 genome sequences in our study separated into three distinct phylogenetic clades. Clade 1 was most closely related to viral sequences from New York, and showed evidence of rapid expansion across the US, while Clade 3 was most closely related to those in Washington state. Clade 2 was localized primarily to the Chicago area with limited evidence of expansion elsewhere. At the time of diagnosis, patients infected with Clade 1 viruses had significantly higher average viral loads in their upper airways relative to patients infected with Clade 2 viruses, independent of time to symptom onset and disease severity.
Interpretation: These results show that multiple variants of SARS-CoV-2 are circulating in the Chicago area that differ in their relative viral loads in patient upper airways. These data suggest that differences in virus genotype impact viral load and may in turn influence viral transmission and spread.
Funding: Dixon Family Translational Research Award, Northwestern University Clinical and Translational Sciences Institute (NUCATS), National Institute of Allergy and Infectious Diseases (NIAID).
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing financial interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
