This is a preprint.
Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater
- PMID: 32511611
- PMCID: PMC7276038
- DOI: 10.1101/2020.04.15.20066746
Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater
Update in
-
Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater.Cell Rep Med. 2020 Sep 22;1(6):100098. doi: 10.1016/j.xcrm.2020.100098. Epub 2020 Aug 31. Cell Rep Med. 2020. PMID: 32904687 Free PMC article.
Abstract
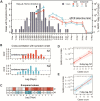
SARS-CoV-2 has recently been detected in feces, which indicates that wastewater may be used to monitor viral prevalence in the community. Here we use RT-qPCR to monitor wastewater for SARS-CoV-2 RNA over a 52-day time course. We show that changes in SARS-CoV-2 RNA concentrations correlate with local COVID-19 epidemiological data (R2=0.9), though detection in wastewater trails symptom onset dates by 5-8 days. We determine a near complete (98.5%) SARS-CoV-2 genome sequence from the wastewater and use phylogenic analysis to infer viral ancestry. Collectively, this work demonstrates how wastewater can be used as a proxy to monitor viral prevalence in the community and how genome sequencing can be used for high-resolution genotyping of the predominant strains circulating in a community.
Conflict of interest statement
Declaration of interests B.W. is the founder of SurGene, LLC, and is an inventor on patent applications related to CRISPR-Cas systems and applications thereof.
Figures

References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., et al. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 728, 138764. - PMC - PubMed
-
- Cheung K.S., Hung I.F., Chan P.P., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W., Tam A.R., et al. (2020). Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples from the Hong Kong Cohort and Systematic Review and Meta-analysis. Gastroenterology. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
