The Expression, Functions and Mechanisms of Circular RNAs in Gynecological Cancers
- PMID: 32512912
- PMCID: PMC7352180
- DOI: 10.3390/cancers12061472
The Expression, Functions and Mechanisms of Circular RNAs in Gynecological Cancers
Abstract
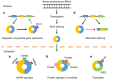
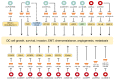
Circular RNAs (circRNAs) are covalently closed, endogenous non-coding RNAs and certain circRNAs are linked to human tumors. Owing to their circular form, circRNAs are protected from degradation by exonucleases, and therefore, they are more stable than linear RNAs. Many circRNAs have been shown to sponge microRNAs, interact with RNA-binding proteins, regulate gene transcription, and be translated into proteins. Mounting evidence suggests that circRNAs are dysregulated in cancer tissues and can mediate various signaling pathways, thus affecting tumorigenesis, metastasis, and remodeling of the tumor microenvironment. First, we review the characteristics, biogenesis, and biological functions of circRNAs, and describe various mechanistic models of circRNAs. Then, we provide a systematic overview of the functional roles of circRNAs in gynecological cancers. Finally, we describe the potential future applications of circRNAs as biomarkers for prognostic stratification and as therapeutic targets in gynecological cancers. Although the function of most circRNAs remains elusive, some individual circRNAs have biologically relevant functions in cervical cancer, ovarian cancer, and endometrial cancer. Certain circRNAs have the potential to serve as biomarkers and therapeutic targets in gynecological cancers.
Keywords: biomarker; cancer diagnosis; cancer treatment; circular RNA; gynecological cancer; non-coding RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A Comprehensive Overview of circRNAs: Emerging Biomarkers and Potential Therapeutics in Gynecological Cancers.Front Cell Dev Biol. 2021 Jul 21;9:709512. doi: 10.3389/fcell.2021.709512. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34368160 Free PMC article. Review.
-
Review on circular RNAs and new insights into their roles in cancer.Comput Struct Biotechnol J. 2021 Jan 22;19:910-928. doi: 10.1016/j.csbj.2021.01.018. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33598105 Free PMC article. Review.
-
Circular RNAs play roles in regulatory networks of cell signaling pathways in human cancers.Life Sci. 2022 Nov 15;309:120975. doi: 10.1016/j.lfs.2022.120975. Epub 2022 Sep 17. Life Sci. 2022. PMID: 36126723 Review.
-
Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers.Pharmacol Ther. 2021 Apr;220:107715. doi: 10.1016/j.pharmthera.2020.107715. Epub 2020 Oct 24. Pharmacol Ther. 2021. PMID: 33141028 Free PMC article. Review.
-
Recent progress in circular RNAs in human cancers.Cancer Lett. 2017 Sep 28;404:8-18. doi: 10.1016/j.canlet.2017.07.002. Epub 2017 Jul 11. Cancer Lett. 2017. PMID: 28705773
Cited by
-
CircHIPK2 Contributes to DDP Resistance and Malignant Behaviors of DDP-Resistant Ovarian Cancer Cells Both in vitro and in vivo Through circHIPK2/miR-338-3p/CHTOP ceRNA Pathway.Onco Targets Ther. 2021 May 13;14:3151-3165. doi: 10.2147/OTT.S291823. eCollection 2021. Onco Targets Ther. 2021. PMID: 34012271 Free PMC article.
-
CircRNA circ_0006677 Inhibits the Progression and Glycolysis in Non-Small-Cell Lung Cancer by Sponging miR-578 and Regulating SOCS2 Expression.Front Pharmacol. 2021 May 13;12:657053. doi: 10.3389/fphar.2021.657053. eCollection 2021. Front Pharmacol. 2021. PMID: 34054537 Free PMC article.
-
Emerging roles of circular RNAs in systemic lupus erythematosus.Mol Ther Nucleic Acids. 2021 Mar 1;24:212-222. doi: 10.1016/j.omtn.2021.02.028. eCollection 2021 Jun 4. Mol Ther Nucleic Acids. 2021. PMID: 33767917 Free PMC article. Review.
-
Circular RNA circRIMS1 Acts as a Sponge of miR-433-3p to Promote Bladder Cancer Progression by Regulating CCAR1 Expression.Mol Ther Nucleic Acids. 2020 Oct 10;22:815-831. doi: 10.1016/j.omtn.2020.10.003. eCollection 2020 Dec 4. Mol Ther Nucleic Acids. 2020. PMID: 33230478 Free PMC article.
-
Silencing of circRNA circ_0001666 Represses EMT in Pancreatic Cancer Through Upregulating miR-1251 and Downregulating SOX4.Front Mol Biosci. 2021 May 13;8:684866. doi: 10.3389/fmolb.2021.684866. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055896 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

