Analysis of 1321 Eubacterium rectale genomes from metagenomes uncovers complex phylogeographic population structure and subspecies functional adaptations
- PMID: 32513234
- PMCID: PMC7278147
- DOI: 10.1186/s13059-020-02042-y
Analysis of 1321 Eubacterium rectale genomes from metagenomes uncovers complex phylogeographic population structure and subspecies functional adaptations
Abstract
Background: Eubacterium rectale is one of the most prevalent human gut bacteria, but its diversity and population genetics are not well understood because large-scale whole-genome investigations of this microbe have not been carried out.
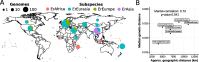
Results: Here, we leverage metagenomic assembly followed by a reference-based binning strategy to screen over 6500 gut metagenomes spanning geography and lifestyle and reconstruct over 1300 E. rectale high-quality genomes from metagenomes. We extend previous results of biogeographic stratification, identifying a new subspecies predominantly found in African individuals and showing that closely related non-human primates do not harbor E. rectale. Comparison of pairwise genetic and geographic distances between subspecies suggests that isolation by distance and co-dispersal with human populations might have contributed to shaping the contemporary population structure of E. rectale. We confirm that a relatively recently diverged E. rectale subspecies specific to Europe consistently lacks motility operons and that it is immotile in vitro, probably due to ancestral genetic loss. The same subspecies exhibits expansion of its carbohydrate metabolism gene repertoire including the acquisition of a genomic island strongly enriched in glycosyltransferase genes involved in exopolysaccharide synthesis.
Conclusions: Our study provides new insights into the population structure and ecology of E. rectale and shows that shotgun metagenomes can enable population genomics studies of microbiota members at a resolution and scale previously attainable only by extensive isolate sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Duncan SH, Flint HJ. Proposal of a neotype strain (A1-86) for Eubacterium rectale. Request for an opinion. Int J Syst Evol Microbiol. 2008;58:1735–1736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

