GPCR-dependent biasing of GIRK channel signaling dynamics by RGS6 in mouse sinoatrial nodal cells
- PMID: 32513692
- PMCID: PMC7322085
- DOI: 10.1073/pnas.2001270117
GPCR-dependent biasing of GIRK channel signaling dynamics by RGS6 in mouse sinoatrial nodal cells
Abstract
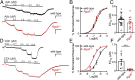
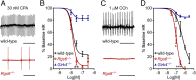
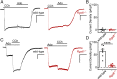
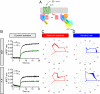
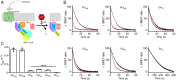
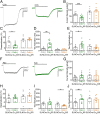
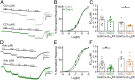
How G protein-coupled receptors (GPCRs) evoke specific biological outcomes while utilizing a limited array of G proteins and effectors is poorly understood, particularly in native cell systems. Here, we examined signaling evoked by muscarinic (M2R) and adenosine (A1R) receptor activation in the mouse sinoatrial node (SAN), the cardiac pacemaker. M2R and A1R activate a shared pool of cardiac G protein-gated inwardly rectifying K+ (GIRK) channels in SAN cells from adult mice, but A1R-GIRK responses are smaller and slower than M2R-GIRK responses. Recordings from mice lacking Regulator of G protein Signaling 6 (RGS6) revealed that RGS6 exerts a GPCR-dependent influence on GIRK-dependent signaling in SAN cells, suppressing M2R-GIRK coupling efficiency and kinetics and A1R-GIRK signaling amplitude. Fast kinetic bioluminescence resonance energy transfer assays in transfected HEK cells showed that RGS6 prefers Gαo over Gαi as a substrate for its catalytic activity and that M2R signals preferentially via Gαo, while A1R does not discriminate between inhibitory G protein isoforms. The impact of atrial/SAN-selective ablation of Gαo or Gαi2 was consistent with these findings. Gαi2 ablation had minimal impact on M2R-GIRK and A1R-GIRK signaling in SAN cells. In contrast, Gαo ablation decreased the amplitude and slowed the kinetics of M2R-GIRK responses, while enhancing the sensitivity and prolonging the deactivation rate of A1R-GIRK signaling. Collectively, our data show that differences in GPCR-G protein coupling preferences, and the Gαo substrate preference of RGS6, shape A1R- and M2R-GIRK signaling dynamics in mouse SAN cells.
Keywords: G protein; Kir3; adenosine; heart rate; muscarinic.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Dyavanapalli J., Dergacheva O., Wang X., Mendelowitz D., Parasympathetic vagal control of cardiac function. Curr. Hypertens. Rep. 18, 22 (2016). - PubMed
-
- Dhein S., van Koppen C. J., Brodde O. E., Muscarinic receptors in the mammalian heart. Pharmacol. Res. 44, 161–182 (2001). - PubMed
-
- Sakmann B., Noma A., Trautwein W., Acetylcholine activation of single muscarinic K+ channels in isolated pacemaker cells of the mammalian heart. Nature 303, 250–253 (1983). - PubMed
-
- Logothetis D. E., Kurachi Y., Galper J., Neer E. J., Clapham D. E., The β γ subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature 325, 321–326 (1987). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

