RIG-I regulates myeloid differentiation by promoting TRIM25-mediated ISGylation
- PMID: 32513696
- PMCID: PMC7322067
- DOI: 10.1073/pnas.1918596117
RIG-I regulates myeloid differentiation by promoting TRIM25-mediated ISGylation
Abstract
Retinoic acid-inducible gene I (RIG-I) is up-regulated during granulocytic differentiation of acute promyelocytic leukemia (APL) cells induced by all-trans retinoic acid (ATRA). It has been reported that RIG-I recognizes virus-specific 5'-ppp-double-stranded RNA (dsRNA) and activates the type I interferons signaling pathways in innate immunity. However, the functions of RIG-I in hematopoiesis remain unclear, especially regarding its possible interaction with endogenous RNAs and the associated pathways that could contribute to the cellular differentiation and maturation. Herein, we identified a number of RIG-I-binding endogenous RNAs in APL cells following ATRA treatment, including the tripartite motif-containing protein 25 (TRIM25) messenger RNA (mRNA). TRIM25 encodes the protein known as an E3 ligase for ubiquitin/interferon (IFN)-induced 15-kDa protein (ISG15) that is involved in RIG-I-mediated antiviral signaling. We show that RIG-I could bind TRIM25 mRNA via its helicase domain and C-terminal regulatory domain, enhancing the stability of TRIM25 transcripts. RIG-I could increase the transcriptional expression of TRIM25 by caspase recruitment domain (CARD) domain through an IFN-stimulated response element. In addition, RIG-I activated other key genes in the ISGylation pathway by activating signal transducer and activator of transcription 1 (STAT1), including the modifier ISG15 and several enzymes responsible for the conjugation of ISG15 to protein substrates. RIG-I cooperated with STAT1/2 and interferon regulatory factor 1 (IRF1) to promote the activation of the ISGylation pathway. The integrity of ISGylation in ATRA or RIG-I-induced cell differentiation was essential given that knockdown of TRIM25 or ISG15 resulted in significant inhibition of this process. Our results provide insight into the role of the RIG-I-TRIM25-ISGylation axis in myeloid differentiation.
Keywords: ISGylation; RIG-I; TRIM25; acute promyelocytic leukemia (APL); myeloid differentiation.
Conflict of interest statement
The authors declare no competing interest.
Figures
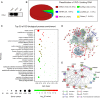

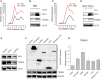
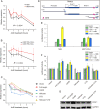
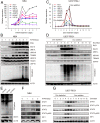
References
-
- Liu T. X. et al. ., Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood 96, 1496–1504 (2000). - PubMed
-
- Hornung V. et al. ., 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 (2006). - PubMed
-
- Yoneyama M. et al. ., The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004). - PubMed
-
- Gack M. U. et al. ., TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

