Analysis of Global Methylome and Gene Expression during Carbon Reserve Mobilization in Stems under Soil Drying
- PMID: 32513834
- PMCID: PMC7401115
- DOI: 10.1104/pp.20.00141
Analysis of Global Methylome and Gene Expression during Carbon Reserve Mobilization in Stems under Soil Drying
Abstract
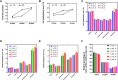
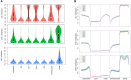
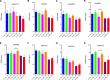
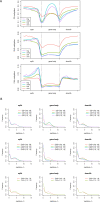
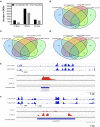
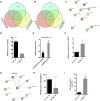
In rice (Oryza sativa), a specific temporary source organ, the stem, is important for grain filling, and moderate soil drying (MD) enhanced carbon reserve flow from stems to increase grain yield. The dynamics and biological relevance of DNA methylation in carbon reserve remobilization during grain filling are unknown. Here, we generated whole-genome single-base resolution maps of the DNA methylome in the stem. During grain filling under MD, we observed an increase in DNA methylation of total cytosines, with more hypomethylated than hypermethylated regions. Genes responsible for DNA methylation and demethylation were up-regulated, suggesting that DNA methylation changes in the stem were regulated by antagonism between DNA methylation and demethylation activity. In addition, methylation in the CG and CHG contexts was negatively associated with gene expression, while that in the CHH context was positively associated with gene expression. A hypermethylated/up-regulated transcription factor of MYBS2 inhibited MYB30 expression and possibly enhanced β-Amylase5 expression, promoting subsequent starch degradation in rice stems under MD treatment. In addition, a hypermethylated/down-regulated transcription factor of ERF24 was predicted to interact with, and thereby decrease the expression of, abscisic acid 8'-hydroxylase1, thus increasing abscisic acid concentration under MD treatment. Our findings provide insight into the DNA methylation dynamics in carbon reserve remobilization of rice stems, demonstrate that MD increased this remobilization, and suggest a link between DNA methylation and gene expression in rice stems during grain filling.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Chan SWL, Henderson IR, Jacobsen SE(2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 - PubMed
-
- Chen HJ, Wang SJ(2012) Abscisic acid enhances starch degradation and sugar transport in rice upper leaf sheaths at the post-heading stage. Acta Physiol Plant 34: 1493–1500
-
- Chen X, Zhou DX(2013) Rice epigenomics and epigenetics: Challenges and opportunities. Curr Opin Plant Biol 16: 164–169 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

