Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits
- PMID: 32513867
- PMCID: PMC7286538
- DOI: 10.1126/scitranslmed.abc3539
Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits
Abstract
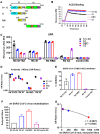
Multiple vaccine candidates against SARS-CoV-2 based on viral spike protein are under development. However, there is limited information on the quality of antibody responses generated with these vaccine modalities. To better understand antibody responses induced by spike protein-based vaccines, we performed a qualitative study by immunizing rabbits with various SARS-CoV-2 spike protein antigens: S ectodomain (S1+S2; amino acids 16 to 1213), which lacks the cytoplasmic and transmembrane domains (CT-TM), the S1 domain (amino acids 16 to 685), the receptor binding domain (RBD) (amino acids 319 to 541), and the S2 domain (amino acids 686 to 1213, lacking the RBD, as control). Resulting antibody quality and function were analyzed by enzyme-linked immunosorbent assay (ELISA), RBD competition assay, surface plasmon resonance (SPR) against different spike proteins in native conformation, and neutralization assays. All three antigens (S1+S2 ectodomain, S1 domain, and RBD), but not S2, generated strong neutralizing antibodies against SARS-CoV-2. Vaccination-induced antibody repertoire was analyzed by SARS-CoV-2 spike genome fragment phage display libraries (SARS-CoV-2 GFPDL), which identified immunodominant epitopes in the S1, S1-RBD, and S2 domains. Furthermore, these analyses demonstrated that the RBD immunogen elicited a higher antibody titer with five-fold higher affinity antibodies to native spike antigens compared with other spike antigens, and antibody affinity correlated strongly with neutralization titers. These findings may help guide rational vaccine design and facilitate development and evaluation of effective therapeutics and vaccines against COVID-19 disease.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

References
-
- John Hopkins University & Medicine Center for Systems Science and Engineering (CSSE), (2020), vol. 2020.
-
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A. S., Liu D., Qin C., Jiang C., Penninger J. M., A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879 (2005). 10.1038/nm1267 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

