Toll-Like Receptors Induce Signal-Specific Reprogramming of the Macrophage Lipidome
- PMID: 32516576
- PMCID: PMC7891175
- DOI: 10.1016/j.cmet.2020.05.003
Toll-Like Receptors Induce Signal-Specific Reprogramming of the Macrophage Lipidome
Abstract
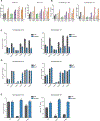
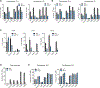
Macrophages reprogram their lipid metabolism in response to activation signals. However, a systems-level understanding of how different pro-inflammatory stimuli reshape the macrophage lipidome is lacking. Here, we use complementary "shotgun" and isotope tracer mass spectrometry approaches to define the changes in lipid biosynthesis, import, and composition of macrophages induced by various Toll-like receptors (TLRs) and inflammatory cytokines. "Shotgun" lipidomics data revealed that different TLRs and cytokines induce macrophages to acquire distinct lipidomes, indicating their specificity in reshaping lipid composition. Mechanistic studies showed that differential reprogramming of lipid composition is mediated by the opposing effects of MyD88- and TRIF-interferon-signaling pathways. Finally, we applied these insights to show that perturbing reprogramming of lipid composition can enhance inflammation and promote host defense to bacterial challenge. These studies provide a framework for understanding how inflammatory stimuli reprogram lipid composition of macrophages while providing a knowledge platform to exploit differential lipidomics to influence immunity.
Keywords: MyD88; acetylated-LDL; host defense; inflammation; interferon; lipidomics; macrophages; stable isotope tracer analysis; stearoyl-CoA desaturase; toll-like receptors.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

