The Dynamic Life of Virus Capsids
- PMID: 32516952
- PMCID: PMC7354500
- DOI: 10.3390/v12060618
The Dynamic Life of Virus Capsids
Abstract
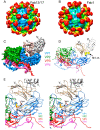
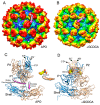
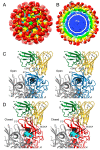
Protein-shelled viruses have been thought as "tin cans" that merely carry the genomic cargo from cell to cell. However, through the years, it has become clear that viruses such as rhinoviruses and caliciviruses are active and dynamic structures waiting for the right environmental cues to deliver their genomic payload to the host cell. In the case of human rhinoviruses, the capsid has empty cavities that decrease the energy required to cause conformational changes, resulting in the capsids "breathing", waiting for the moment when the receptor binds for it to release its genome. Most strikingly, the buried N-termini of VP1 and VP4 are transiently exposed during this process. A more recent example of a "living" protein capsid is mouse norovirus (MNV). This family of viruses have a large protruding (P) domain that is loosely attached to the shell via a single-polypeptide tether. Small molecules found in the gut, such as bile salts, cause the P domains to rotate and collapse onto the shell surface. Concomitantly, bile alters the conformation of the P domain itself from one that binds antibodies to one that recognizes receptors. In this way, MNV appears to use capsid flexibility to present one face to the immune system and a completely different one to attack the host tissue. Therefore, it appears that even protein-shelled viruses have developed an impressive array of tricks to dodge our immune system and efficiently attack the host.
Keywords: antibodies; flexibility; norovirus; rhinovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Rueckert R.R. Picornaviridae and Their Replication. In: Fields B.N., Knipe D.M., editors. Fundamental Virology. Raven Press; New York, NY, USA: 1996. pp. 609–654.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

