Insights into the Role of Estrogen Receptor β in Triple-Negative Breast Cancer
- PMID: 32516978
- PMCID: PMC7353068
- DOI: 10.3390/cancers12061477
Insights into the Role of Estrogen Receptor β in Triple-Negative Breast Cancer
Abstract
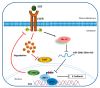
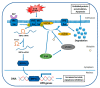
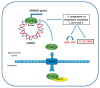
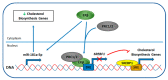
Estrogen receptors (ERα and ERβ) are ligand-activated transcription factors that play different roles in gene regulation and show both overlapping and specific tissue distribution patterns. ERβ, contrary to the oncogenic ERα, has been shown to act as an oncosuppressor in several instances. However, while the tumor-promoting actions of ERα are well-known, the exact role of ERβ in carcinogenesis and tumor progression is not yet fully understood. Indeed, to date, highly variable and even opposite effects have been ascribed to ERβ in cancer, including for example both proliferative and growth-inhibitory actions. Recently ERβ has been proposed as a potential target for cancer therapy, since it is expressed in a variety of breast cancers (BCs), including triple-negative ones (TNBCs). Because of the dependence of TNBCs on active cellular signaling, numerous studies have attempted to unravel the mechanism(s) behind ERβ-regulated gene expression programs but the scenario has not been fully revealed. We comprehensively reviewed the current state of knowledge concerning ERβ role in TNBC biology, focusing on the different signaling pathways and cellular processes regulated by this transcription factor, as they could be useful in identifying new diagnostic and therapeutic approaches for TNBC.
Keywords: TNBC; cancer cell metabolism; estrogen receptor β; oncosuppressor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

