Evolution of the T-Cell Receptor (TR) Loci in the Adaptive Immune Response: The Tale of the TRG Locus in Mammals
- PMID: 32517024
- PMCID: PMC7349638
- DOI: 10.3390/genes11060624
Evolution of the T-Cell Receptor (TR) Loci in the Adaptive Immune Response: The Tale of the TRG Locus in Mammals
Abstract
T lymphocytes are the principal actors of vertebrates' cell-mediated immunity. Like B cells, they can recognize an unlimited number of foreign molecules through their antigen-specific heterodimer receptors (TRs), which consist of αβ or γδ chains. The diversity of the TRs is mainly due to the unique organization of the genes encoding the α, β, γ, and δ chains. For each chain, multi-gene families are arranged in a TR locus, and their expression is guaranteed by the somatic recombination process. A great plasticity of the gene organization within the TR loci exists among species. Marked structural differences affect the TR γ (TRG) locus. The recent sequencing of multiple whole genome provides an opportunity to examine the TR gene repertoire in a systematic and consistent fashion. In this review, we report the most recent findings on the genomic organization of TRG loci in mammalian species in order to show differences and similarities. The comparison revealed remarkable diversification of both the genomic organization and gene repertoire across species, but also unexpected evolutionary conservation, which highlights the important role of the T cells in the immune response.
Keywords: IMGT; T cell receptor; TRG genes; TRG locus; evolution; immunogenomics; mammals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

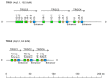
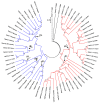
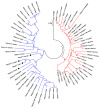

References
-
- Lefranc M.-P., Lefranc G. The T Cell Receptor FactsBook. Academic Press. Harcourt Science and Technology Company; San Diego, CA, USA: 2001.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

