Oncogenic Properties of the EBV ZEBRA Protein
- PMID: 32517128
- PMCID: PMC7352903
- DOI: 10.3390/cancers12061479
Oncogenic Properties of the EBV ZEBRA Protein
Abstract
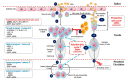
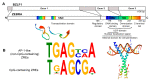
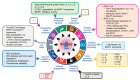
Epstein Barr Virus (EBV) is one of the most common human herpesviruses. After primary infection, it can persist in the host throughout their lifetime in a latent form, from which it can reactivate following specific stimuli. EBV reactivation is triggered by transcriptional transactivator proteins ZEBRA (also known as Z, EB-1, Zta or BZLF1) and RTA (also known as BRLF1). Here we discuss the structural and functional features of ZEBRA, its role in oncogenesis and its possible implication as a prognostic or diagnostic marker. Modulation of host gene expression by ZEBRA can deregulate the immune surveillance, allow the immune escape, and favor tumor progression. It also interacts with host proteins, thereby modifying their functions. ZEBRA is released into the bloodstream by infected cells and can potentially penetrate any cell through its cell-penetrating domain; therefore, it can also change the fate of non-infected cells. The features of ZEBRA described in this review outline its importance in EBV-related malignancies.
Keywords: BZLF1; EBV; ZEBRA; Zta; lytic cycle; oncogenesis; transactivation; transcription; viral-host interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- IARC Proceedings of the IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Epstein-Barr Virus and Kaposi’s Sarcoma Herpesvirus/Human Herpesvirus 8. Lyon, France, 17-24 June 1997. IARC Monogr. Eval. Carcinog. Risks Hum. 1997;70:1–492. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

