Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis
- PMID: 32517243
- PMCID: PMC7356249
- DOI: 10.3390/biom10060874
Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis
Abstract
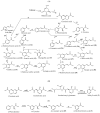
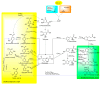
Biotechnological production of phenolic acids is attracting increased interest due to their superior antioxidant activity, as well as other antimicrobial, dietary, and health benefits. As secondary metabolites, primarily found in plants and fungi, they are effective free radical scavengers due to the phenolic group available in their structure. Therefore, phenolic acids are widely utilised by pharmaceutical, food, cosmetic, and chemical industries. A demand for phenolic acids is mostly satisfied by utilising chemically synthesised compounds, with only a low quantity obtained from natural sources. As an alternative to chemical synthesis, environmentally friendly bio-based technologies are necessary for development in large-scale production. One of the most promising sustainable technologies is the utilisation of microbial cell factories for biosynthesis of phenolic acids. In this paper, we perform a systematic comparison of the best known natural sources of phenolic acids. The advances and prospects in the development of microbial cell factories for biosynthesis of these bioactive compounds are discussed in more detail. A special consideration is given to the modern production methods and analytics of phenolic acids.
Keywords: analytical methods; antioxidant activity; biorefinery; biosensor; extraction; metabolic engineering; microbial fermentation; phenolic acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Barros J., Escamilla-Trevino L., Song L., Rao X., Serrani-Yarce J.C., Palacios M.D., Engle N., Choudhury F.K., Tschaplinski T.J., Venables B.J., et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019;10:1994. doi: 10.1038/s41467-019-10082-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

