Collapsing the list of myocardial infarction-related differentially expressed genes into a diagnostic signature
- PMID: 32517814
- PMCID: PMC7285786
- DOI: 10.1186/s12967-020-02400-1
Collapsing the list of myocardial infarction-related differentially expressed genes into a diagnostic signature
Abstract
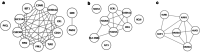
Background: Myocardial infarction (MI) is one of the most severe manifestations of coronary artery disease (CAD) and the leading cause of death from non-infectious diseases worldwide. It is known that the central component of CAD pathogenesis is a chronic vascular inflammation. However, the mechanisms underlying the changes that occur in T, B and NK lymphocytes, monocytes and other immune cells during CAD and MI are still poorly understood. One of those pathogenic mechanisms might be the dysregulation of intracellular signaling pathways in the immune cells.
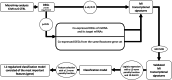
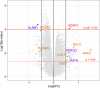
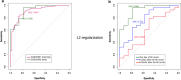
Methods: In the present study we performed a transcriptome profiling in peripheral blood mononuclear cells of MI patients and controls. The machine learning algorithm was then used to search for MI-associated signatures, that could reflect the dysregulation of intracellular signaling pathways.
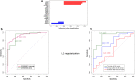
Results: The genes ADAP2, KLRC1, MIR21, PDGFD and CD14 were identified as the most important signatures for the classification model with L1-norm penalty function. The classifier output quality was equal to 0.911 by Receiver Operating Characteristic metric on test data. These results were validated on two independent open GEO datasets. Identified MI-associated signatures can be further assisted in MI diagnosis and/or prognosis.
Conclusions: Thus, our study presents a pipeline for collapsing the list of differential expressed genes, identified by high-throughput techniques, in order to define disease-associated diagnostic signatures.
Keywords: Machine learning; Myocardial infarction; Transcriptional signatures; Transcriptomics; miRNA.
Conflict of interest statement
The authors declare that they have no conflict of interest relating to the conduct of this study or the publication of this manuscript.
Figures
Similar articles
-
Unlocking the potential of microRNAs: machine learning identifies key biomarkers for myocardial infarction diagnosis.Cardiovasc Diabetol. 2023 Sep 11;22(1):247. doi: 10.1186/s12933-023-01957-7. Cardiovasc Diabetol. 2023. PMID: 37697288 Free PMC article.
-
Genome-wide transcriptome study using deep RNA sequencing for myocardial infarction and coronary artery calcification.BMC Med Genomics. 2021 Feb 10;14(1):45. doi: 10.1186/s12920-020-00838-2. BMC Med Genomics. 2021. PMID: 33568140 Free PMC article.
-
Characterization of Systemic and Culprit-Coronary Artery miR-483-5p Expression in Chronic CAD and Acute Myocardial Infarction Male Patients.Int J Mol Sci. 2023 May 10;24(10):8551. doi: 10.3390/ijms24108551. Int J Mol Sci. 2023. PMID: 37239897 Free PMC article.
-
Identification of gene expression profiles in myocardial infarction: a systematic review and meta-analysis.BMC Med Genomics. 2018 Nov 27;11(1):109. doi: 10.1186/s12920-018-0427-x. BMC Med Genomics. 2018. PMID: 30482209 Free PMC article.
-
MicroRNAs expression profiles in cardiovascular diseases.Biomed Res Int. 2014;2014:985408. doi: 10.1155/2014/985408. Epub 2014 Jun 12. Biomed Res Int. 2014. PMID: 25013816 Free PMC article. Review.
Cited by
-
Impact of medication on blood transcriptome reveals off-target regulations of beta-blockers.PLoS One. 2022 Apr 21;17(4):e0266897. doi: 10.1371/journal.pone.0266897. eCollection 2022. PLoS One. 2022. PMID: 35446883 Free PMC article.
-
Integrated whole-genome gene expression analysis reveals an atlas of dynamic immune landscapes after myocardial infarction.Front Cardiovasc Med. 2023 Mar 3;10:1087721. doi: 10.3389/fcvm.2023.1087721. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 36937942 Free PMC article.
-
Atherosclerotic plaque vulnerability quantification system for clinical and biological interpretability.iScience. 2023 Aug 9;26(9):107587. doi: 10.1016/j.isci.2023.107587. eCollection 2023 Sep 15. iScience. 2023. PMID: 37664595 Free PMC article.
-
Identification through machine learning of potential immune- related gene biomarkers associated with immune cell infiltration in myocardial infarction.BMC Cardiovasc Disord. 2023 Mar 28;23(1):163. doi: 10.1186/s12872-023-03196-w. BMC Cardiovasc Disord. 2023. PMID: 36978012 Free PMC article.
-
RPS27A as a potential clock-related diagnostic biomarker for myocardial infarction: Comprehensive bioinformatics analysis and experimental validation.Clinics (Sao Paulo). 2025 May 22;80:100677. doi: 10.1016/j.clinsp.2025.100677. eCollection 2025. Clinics (Sao Paulo). 2025. PMID: 40409241 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

