Endothelial-to-mesenchymal transition compromises vascular integrity to induce Myc-mediated metabolic reprogramming in kidney fibrosis
- PMID: 32518142
- PMCID: PMC7790440
- DOI: 10.1126/scisignal.aaz2597
Endothelial-to-mesenchymal transition compromises vascular integrity to induce Myc-mediated metabolic reprogramming in kidney fibrosis
Abstract
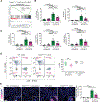
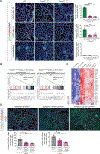
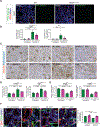
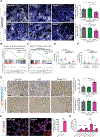
Endothelial-to-mesenchymal transition (EndMT) is a cellular transdifferentiation program in which endothelial cells partially lose their endothelial identity and acquire mesenchymal-like features. Renal capillary endothelial cells can undergo EndMT in association with persistent damage of the renal parenchyma. The functional consequence(s) of EndMT in kidney fibrosis remains unexplored. Here, we studied the effect of Twist or Snail deficiency in endothelial cells on EndMT in kidney fibrosis. Conditional deletion of Twist1 (which encodes Twist) or Snai1 (which encodes Snail) in VE-cadherin+ or Tie1+ endothelial cells inhibited the emergence of EndMT and improved kidney fibrosis in two different kidney injury/fibrosis mouse models. Suppression of EndMT limited peritubular vascular leakage, reduced tissue hypoxia, and preserved tubular epithelial health and function. Hypoxia, which was exacerbated by EndMT, resulted in increased Myc abundance in tubular epithelial cells, enhanced glycolysis, and suppression of fatty acid oxidation. Pharmacological suppression or epithelial-specific genetic ablation of Myc in tubular epithelial cells ameliorated fibrosis and restored renal parenchymal function and metabolic homeostasis. Together, these findings demonstrate a functional role for EndMT in the response to kidney capillary endothelial injury and highlight the contribution of endothelial-epithelial cross-talk in the development of kidney fibrosis with a potential for therapeutic intervention.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



Comment in
-
The interdependence of renal epithelial and endothelial metabolism and cell state.Sci Signal. 2020 Jun 9;13(635):eabb8834. doi: 10.1126/scisignal.abb8834. Sci Signal. 2020. PMID: 32518144 Free PMC article.
References
-
- Eisenberg LM and Markwald RR, Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ Res, 1995. 77(1): p. 1–6. - PubMed
-
- Zeisberg EM, et al., Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med, 2007. 13(8): p. 952–61. - PubMed
-
- Zeisberg EM, et al., Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res, 2007. 67(21): p. 10123–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

