Nitrogen Source Governs Community Carbon Metabolism in a Model Hypersaline Benthic Phototrophic Biofilm
- PMID: 32518194
- PMCID: PMC7289588
- DOI: 10.1128/mSystems.00260-20
Nitrogen Source Governs Community Carbon Metabolism in a Model Hypersaline Benthic Phototrophic Biofilm
Abstract
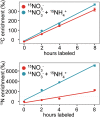
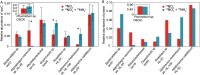
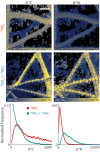
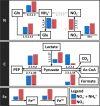
Increasing anthropogenic inputs of fixed nitrogen are leading to greater eutrophication of aquatic environments, but it is unclear how this impacts the flux and fate of carbon in lacustrine and riverine systems. Here, we present evidence that the form of nitrogen governs the partitioning of carbon among members in a genome-sequenced, model phototrophic biofilm of 20 members. Consumption of NO3 - as the sole nitrogen source unexpectedly resulted in more rapid transfer of carbon to heterotrophs than when NH4 + was also provided, suggesting alterations in the form of carbon exchanged. The form of nitrogen dramatically impacted net community nitrogen, but not carbon, uptake rates. Furthermore, this alteration in nitrogen form caused very large but focused alterations to community structure, strongly impacting the abundance of only two species within the biofilm and modestly impacting a third member species. Our data suggest that nitrogen metabolism may coordinate coupled carbon-nitrogen biogeochemical cycling in benthic biofilms and, potentially, in phototroph-heterotroph consortia more broadly. It further indicates that the form of nitrogen inputs may significantly impact the contribution of these communities to carbon partitioning across the terrestrial-aquatic interface.IMPORTANCE Anthropogenic inputs of nitrogen into aquatic ecosystems, and especially those of agricultural origin, involve a mix of chemical species. Although it is well-known in general that nitrogen eutrophication markedly influences the metabolism of aquatic phototrophic communities, relatively little is known regarding whether the specific chemical form of nitrogen inputs matter. Our data suggest that the nitrogen form alters the rate of nitrogen uptake significantly, whereas corresponding alterations in carbon uptake were minor. However, differences imposed by uptake of divergent nitrogen forms may result in alterations among phototroph-heterotroph interactions that rewire community metabolism. Furthermore, our data hint that availability of other nutrients (i.e., iron) might mediate the linkage between carbon and nitrogen cycling in these communities. Taken together, our data suggest that different nitrogen forms should be examined for divergent impacts on phototrophic communities in fluvial systems and that these anthropogenic nitrogen inputs may significantly differ in their ultimate biogeochemical impacts.
Keywords: carbon cycling; cyanobacteria; mass spectrometry; nitrogen cycling; stable isotopes.
Copyright © 2020 Anderton et al.
Figures
References
-
- Cullen JJ. 1999. Iron, nitrogen and phosphorus in the ocean. Nature 402:372. doi:10.1038/46469. - DOI
-
- Khalili B, Ogunseitan OA, Goulden ML, Allison SD. 2016. Interactive effects of precipitation manipulation and nitrogen addition on soil properties in California grassland and shrubland. Appl Soil Ecol 107:144–153. doi:10.1016/j.apsoil.2016.05.018. - DOI
-
- Zeng J, Liu X, Song L, Lin X, Zhang H, Shen C, Chu H. 2016. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol Biochem 92:41–49. doi:10.1016/j.soilbio.2015.09.018. - DOI
-
- Karl D, Letelier R, Tupas L, Dore J, Christian J, Hebel D. 1997. The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean. Nature 388:533–538. doi:10.1038/41474. - DOI
LinkOut - more resources
Full Text Sources

