The genetic basis of sex determination in grapes
- PMID: 32518223
- PMCID: PMC7283251
- DOI: 10.1038/s41467-020-16700-z
The genetic basis of sex determination in grapes
Abstract
It remains a major challenge to identify the genes and mutations that lead to plant sexual differentiation. Here, we study the structure and evolution of the sex-determining region (SDR) in Vitis species. We report an improved, chromosome-scale Cabernet Sauvignon genome sequence and the phased assembly of nine wild and cultivated grape genomes. By resolving twenty Vitis SDR haplotypes, we compare male, female, and hermaphrodite haplotype structures and identify sex-linked regions. Coupled with gene expression data, we identify a candidate male-sterility mutation in the VviINP1 gene and potential female-sterility function associated with the transcription factor VviYABBY3. Our data suggest that dioecy has been lost during domestication through a rare recombination event between male and female haplotypes. This work significantly advances the understanding of the genetic basis of sex determination in Vitis and provides the information necessary to rapidly identify sex types in grape breeding programs.
Conflict of interest statement
The authors declare no competing interests.
Figures
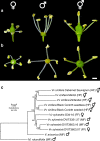
 represent female, male, and hermaphrodite individuals, respectively. Numbers associated with nodes reflect bootstrap values (see “Methods”). Scale bar is in the unit of the number of substitutions per site.
represent female, male, and hermaphrodite individuals, respectively. Numbers associated with nodes reflect bootstrap values (see “Methods”). Scale bar is in the unit of the number of substitutions per site.
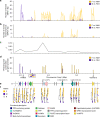



 represent female, male and hermaphrodite individuals, respectively.
represent female, male and hermaphrodite individuals, respectively.References
-
- Westergaard M. The mechanism of sex determination in dioecious flowering plants. Adv. Genet. 1958;9:217–281. - PubMed
-
- Charlesworth, D. in Evolution—essays in honour of John Maynard Smith 237–268 (Cambridge University Press, Cambridge, 1985).
-
- Charlesworth B, Charlesworth DA. Model for the evolution of dioecy and gynodioecy. Am. Nat. 1978;112:975–997.
-
- Charlesworth D. Plant sex chromosomes. Annu. Rev. Plant Biol. 2016;67:397–420. - PubMed
-
- Bull, J. J. Evolution of Sex Determining Mechanism (Benjamin/Cummings, Menlo Park, 1983).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

