A sister lineage of the Mycobacterium tuberculosis complex discovered in the African Great Lakes region
- PMID: 32518235
- PMCID: PMC7283319
- DOI: 10.1038/s41467-020-16626-6
A sister lineage of the Mycobacterium tuberculosis complex discovered in the African Great Lakes region
Abstract
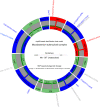
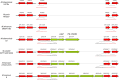
The human- and animal-adapted lineages of the Mycobacterium tuberculosis complex (MTBC) are thought to have expanded from a common progenitor in Africa. However, the molecular events that accompanied this emergence remain largely unknown. Here, we describe two MTBC strains isolated from patients with multidrug resistant tuberculosis, representing an as-yet-unknown lineage, named Lineage 8 (L8), seemingly restricted to the African Great Lakes region. Using genome-based phylogenetic reconstruction, we show that L8 is a sister clade to the known MTBC lineages. Comparison with other complete mycobacterial genomes indicate that the divergence of L8 preceded the loss of the cobF genome region - involved in the cobalamin/vitamin B12 synthesis - and gene interruptions in a subsequent common ancestor shared by all other known MTBC lineages. This discovery further supports an East African origin for the MTBC and provides additional molecular clues on the ancestral genome reduction associated with adaptation to a pathogenic lifestyle.
Conflict of interest statement
P.S. was a consultant of Genoscreen; C.G. and S.D. were employees of the same company. The other authors declare no competing interests.
Figures



References
-
- Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018;16:202–213. - PubMed
-
- World Health Organization. Global Tuberculosis Report2018 (WHO, 2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

