GLOBAL AND TARGETED PROFILING OF GTP-BINDING PROTEINS IN BIOLOGICAL SAMPLES BY MASS SPECTROMETRY
- PMID: 32519381
- PMCID: PMC7725852
- DOI: 10.1002/mas.21637
GLOBAL AND TARGETED PROFILING OF GTP-BINDING PROTEINS IN BIOLOGICAL SAMPLES BY MASS SPECTROMETRY
Abstract
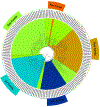
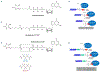
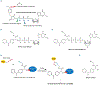
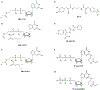
GTP-binding proteins are among the most important enzyme families that are involved in a plethora of biological processes. However, owing to the enormous diversity of the nucleotide-binding protein family, comprehensive analyses of the expression level, structure, activity, and regulatory mechanisms of GTP-binding proteins remain challenging with the use of conventional approaches. The many advances in mass spectrometry (MS) instrumentation and data acquisition methods, together with a variety of enrichment approaches in sample preparation, render MS a powerful tool for the comprehensive characterizations of the activities and expression levels of various GTP-binding proteins. We review herein the recent developments in the application of MS-based techniques, together with general and widely used affinity enrichment approaches, for the proteome-wide and targeted capture, identification, and quantification of GTP-binding proteins. The working principles, advantages, and limitations of various strategies for profiling the expression level, activity, posttranslational modifications, and interactome of GTP-binding proteins are discussed. It can be envisaged that future applications of MS-based proteomics will lead to a better understanding about the roles of GTP-binding proteins in different biological processes and human diseases. © 2020 John Wiley & Sons Ltd. Mass Spec Rev.
Keywords: GTP-binding proteins; activity-based protein profiling; posttranslational modifications; shotgun proteomics; small GTPases; targeted proteomics.
© 2020 John Wiley & Sons Ltd.
Figures



References
-
- Adam GC, Sorensen EJ and Cravatt BF (2002). “Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype.” Nature Biotechnology 20(8): 805–809. - PubMed
-
- Aebersold R and Mann M (2003). “Mass spectrometry-based proteomics.” Nature 422(6928): 198–207. - PubMed
-
- Bagci H, Sriskandarajah N, Robert A, Boulais J, Elkholi IE, Tran V, Lin ZY, Thibault MP, Dube N, Faubert D, Hipfner DR, Gingras AC and Cote JF (2019). “Mapping the proximity interaction network of the Rho-family GTPases reveals signalling pathways and regulatory mechanisms.” Nat Cell Biol 22: 120–134. - PubMed
-
- Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B and Drewes G (2007). “Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors.” Nat Biotechnol 25(9): 1035–1044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

