Movement of St. Louis encephalitis virus in the Western United States, 2014- 2018
- PMID: 32520944
- PMCID: PMC7307790
- DOI: 10.1371/journal.pntd.0008343
Movement of St. Louis encephalitis virus in the Western United States, 2014- 2018
Abstract
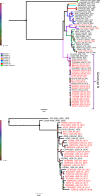
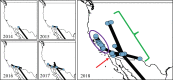
St. Louis encephalitis virus (SLEV) is a flavivirus that circulates in an enzootic cycle between birds and mosquitoes and can also infect humans to cause febrile disease and sometimes encephalitis. Although SLEV is endemic to the United States, no activity was detected in California during the years 2004 through 2014, despite continuous surveillance in mosquitoes and sentinel chickens. In 2015, SLEV-positive mosquito pools were detected in Maricopa County, Arizona, concurrent with an outbreak of human SLEV disease. SLEV-positive mosquito pools were also detected in southeastern California and Nevada in summer 2015. From 2016 to 2018, SLEV was detected in mosquito pools throughout southern and central California, Oregon, Idaho, and Texas. To understand genetic relatedness and geographic dispersal of SLEV in the western United States since 2015, we sequenced four historical genomes (3 from California and 1 from Louisiana) and 26 contemporary SLEV genomes from mosquito pools from locations across the western US. Bayesian phylogeographic approaches were then applied to map the recent spread of SLEV. Three routes of SLEV dispersal in the western United States were identified: Arizona to southern California, Arizona to Central California, and Arizona to all locations east of the Sierra Nevada mountains. Given the topography of the Western United States, these routes may have been limited by mountain ranges that influence the movement of avian reservoirs and mosquito vectors, which probably represents the primary mechanism of SLEV dispersal. Our analysis detected repeated SLEV introductions from Arizona into southern California and limited evidence of year-to-year persistence of genomes of the same ancestry. By contrast, genetic tracing suggests that all SLEV activity since 2015 in central California is the result of a single persistent SLEV introduction. The identification of natural barriers that influence SLEV dispersal enhances our understanding of arbovirus ecology in the western United States and may also support regional public health agencies in implementing more targeted vector mitigation efforts to protect their communities more effectively.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures