IRE1α regulates macrophage polarization, PD-L1 expression, and tumor survival
- PMID: 32520957
- PMCID: PMC7307794
- DOI: 10.1371/journal.pbio.3000687
IRE1α regulates macrophage polarization, PD-L1 expression, and tumor survival
Abstract
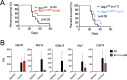
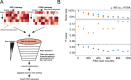
In the tumor microenvironment, local immune dysregulation is driven in part by macrophages and dendritic cells that are polarized to a mixed proinflammatory/immune-suppressive phenotype. The unfolded protein response (UPR) is emerging as the possible origin of these events. Here we report that the inositol-requiring enzyme 1 (IRE1α) branch of the UPR is directly involved in the polarization of macrophages in vitro and in vivo, including the up-regulation of interleukin 6 (IL-6), IL-23, Arginase1, as well as surface expression of CD86 and programmed death ligand 1 (PD-L1). Macrophages in which the IRE1α/X-box binding protein 1 (Xbp1) axis is blocked pharmacologically or deleted genetically have significantly reduced polarization and CD86 and PD-L1 expression, which was induced independent of IFNγ signaling, suggesting a novel mechanism in PD-L1 regulation in macrophages. Mice with IRE1α- but not Xbp1-deficient macrophages showed greater survival than controls when implanted with B16.F10 melanoma cells. Remarkably, we found a significant association between the IRE1α gene signature and CD274 gene expression in tumor-infiltrating macrophages in humans. RNA sequencing (RNASeq) analysis showed that bone marrow-derived macrophages with IRE1α deletion lose the integrity of the gene connectivity characteristic of regulated IRE1α-dependent decay (RIDD) and the ability to activate CD274 gene expression. Thus, the IRE1α/Xbp1 axis drives the polarization of macrophages in the tumor microenvironment initiating a complex immune dysregulation leading to failure of local immune surveillance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

