Chromatin Priming Renders T Cell Tolerance-Associated Genes Sensitive to Activation below the Signaling Threshold for Immune Response Genes
- PMID: 32521273
- PMCID: PMC7296351
- DOI: 10.1016/j.celrep.2020.107748
Chromatin Priming Renders T Cell Tolerance-Associated Genes Sensitive to Activation below the Signaling Threshold for Immune Response Genes
Abstract
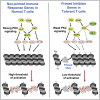
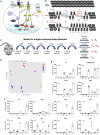
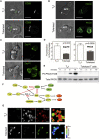
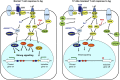
Immunological homeostasis in T cells is maintained by a tightly regulated signaling and transcriptional network. Full engagement of effector T cells occurs only when signaling exceeds a critical threshold that enables induction of immune response genes carrying an epigenetic memory of prior activation. Here we investigate the underlying mechanisms causing the suppression of normal immune responses when T cells are rendered anergic by tolerance induction. By performing an integrated analysis of signaling, epigenetic modifications, and gene expression, we demonstrate that immunological tolerance is established when both signaling to and chromatin priming of immune response genes are weakened. In parallel, chromatin priming of immune-repressive genes becomes boosted, rendering them sensitive to low levels of signaling below the threshold needed to activate immune response genes. Our study reveals how repeated exposure to antigens causes an altered epigenetic state leading to T cell anergy and tolerance, representing a basis for treating auto-immune diseases.
Keywords: CTLA4; Cbl-b; IL-10; T cell; chromatin; epigenetic; gene regulation; signaling; tolerance; transcription factor.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




References
-
- Anderson P.O., Sundstedt A., Yazici Z., Minaee S., O’Neill E.J., Woolf R., Nicolson K., Whitley N., Li L., Li S. IL-2 overcomes the unresponsiveness but fails to reverse the regulatory function of antigen-induced T regulatory cells. J. Immunol. 2005;174:310–319. - PubMed
-
- Bachmaier K., Krawczyk C., Kozieradzki I., Kong Y.Y., Sasaki T., Oliveira-dos-Santos A., Mariathasan S., Bouchard D., Wakeham A., Itie A. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

