High-Throughput Identification of Adapters in Single-Read Sequencing Data
- PMID: 32521604
- PMCID: PMC7356586
- DOI: 10.3390/biom10060878
High-Throughput Identification of Adapters in Single-Read Sequencing Data
Abstract
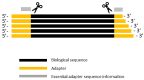
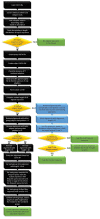
Sequencing datasets available in public repositories are already high in number, and their growth is exponential. Raw sequencing data files constitute a substantial portion of these data, and they need to be pre-processed for any downstream analyses. The removal of adapter sequences is the first essential step. Tools available for the automated detection of adapters in single-read sequencing protocol datasets have certain limitations. To explore these datasets, one needs to retrieve the information on adapter sequences from the methods sections of appropriate research articles. This can be time-consuming in metadata analyses. Moreover, not all research articles provide the information on adapter sequences. We have developed adapt_find, a tool that automates the process of adapter sequences identification in raw single-read sequencing datasets. We have verified adapt_find through testing a number of publicly available datasets. adapt_find secures a robust, reliable and high-throughput process across different sequencing technologies and various adapter designs. It does not need prior knowledge of the adapter sequences. We also produced associated tools: random_mer, for the detection of random N bases either on one or both termini of the reads, and fastqc_parser, for consolidating the results from FASTQC outputs. Together, this is a valuable tool set for metadata analyses on multiple sequencing datasets.
Keywords: 454 pyrosequencing; Illumina; Ion-Torrent; SOLiD; adapter oligonucleotides; adapter trimming; randomized adapters; single-read sequencing; small RNA sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. - DOI
-
- Simon A. FastQC: A Quality Control Tool for High Throughput Sequence Data. [(accessed on 17 March 2020)]; Available online: https://archive.st/archive/2020/3/www.bioinformatics.babraham.ac.uk/4af3....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

