A comprehensive custom panel evaluation for routine hereditary cancer testing: improving the yield of germline mutation detection
- PMID: 32522261
- PMCID: PMC7288470
- DOI: 10.1186/s12967-020-02391-z
A comprehensive custom panel evaluation for routine hereditary cancer testing: improving the yield of germline mutation detection
Abstract
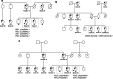
Background: In the context of our Regional Program of Hereditary Cancer, individuals fulfilling the criteria are tested for germline mutations to subsequently establish the clinical management. Our standard diagnostic approach focuses on sequencing a few classic high-risk genes, a method that frequently renders uninformative genetic results. This study aims to examine the improved yield offered by an On-Demand panel.
Methods: We designed an On-Demand panel for the analysis of 35-genes associated with inherited cancer susceptibility in a total of 128 cases of Hereditary Breast and Ovarian Cancer (HBOC) and Hereditary Nonpolyposis Colorectal Cancer (HNPCC).
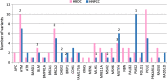
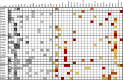
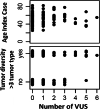
Results: Eighteen deleterious mutations were detected, in both routinely (BRCA2, MLH1, MSH2, PMS2) and non-routinely (ATM, BLM, BRIP1, CHEK2, MUTYH) tested genes. The screening extended to 35 genes rendered by patients carrying several- up to 6-Variants of Unknown Significance (VUS). Moreover, we confirmed the splicing disruption at RNA level for a not previously reported BRIP1 splicing mutation. Using an On-Demand panel, we identified 18 pathogenic mutation carriers, seven of which would have gone unnoticed with traditional analysis.
Conclusions: Our results reinforce the utility of NGS gene panels in the diagnostic routine to increase the performance of genetic testing, especially in individuals from families with overlapping cancer phenotypes.
Keywords: Genetic counselling; Germline mutation; Hereditary Cancer syndrome; Next generation sequencing; On-Demand gene panel.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
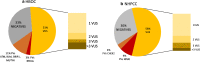
References
-
- Ramírez-Calvo M, García-Casado Z, Fernández-Serra A, de Juan I, Palanca S, Oltra S, et al. Implementation of massive sequencing in the genetic diagnosis of hereditary cancer syndromes: diagnostic performance in the hereditary cancer programme of the valencia community (FamCan-NGS) Hereditary Cancer Clin Pract. 2019;17:3. doi: 10.1186/s13053-019-0104-x. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

