DNA markers based on retrotransposon insertion polymorphisms can detect short DNA fragments for strawberry cultivar identification
- PMID: 32523405
- PMCID: PMC7272250
- DOI: 10.1270/jsbbs.19116
DNA markers based on retrotransposon insertion polymorphisms can detect short DNA fragments for strawberry cultivar identification
Abstract
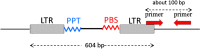
In this study, DNA markers were developed for discrimination of strawberry (Fragaria × ananassa L.) cultivars based on retrotransposon insertion polymorphisms. We performed a comprehensive genomic search to identify retrotransposon insertion sites and subsequently selected one retrotransposon family, designated CL3, which provided reliable discrimination among strawberry cultivars. Through analyses of 75 strawberry cultivars, we developed eight cultivar-specific markers based on CL3 retrotransposon insertion sites. Used in combination with 10 additional polymorphic markers, we differentiated 35 strawberry cultivars commonly cultivated in Japan. In addition, we demonstrated that the retrotransposon-based markers were effective for PCR detection of DNA extracted from processed food materials, whereas a SSR marker was ineffective. These results indicated that the retrotransposon-based markers are useful for cultivar discrimination for processed food products, such as jams, in which DNA may be fragmented or degraded.
Keywords: Fragaria × ananassa; PCR product; high-throughput sequencing; processed foods; retrotransposon insertion polymorphisms.
Copyright © 2020 by JAPANESE SOCIETY OF BREEDING.
Figures



References
-
- Akitake H., Tahara M., Monden Y., Takasaki K. and Futo S. (2013) Strawberry cultivar identification by retrotransposon insertion polymorphisms. DNA Polymorph. 21: 64–72.
-
- Arnau G., Lallemand J. and Bourgoin M. (2003) Fast and reliable strawberry cultivar identification using inter simple sequence repeat (ISSR) amplification. Euphytica 129: 69–79.
-
- Bonoli, M., G. Cipriani, L. Manzecchi and W. Faedi (2005) Utilization of microsatellite molecular markers for characterization of varieties in strawberry [Fragaria Spp.]. Rivista Di Frutticoltura E Di Ortofloricoltura 67.
-
- Degani C., Rowland L.J., Levi A., Hortynski J.A. and Galletta G.J. (1998) DNA fingerprinting of strawberry (Fragaria × ananassa) cultivars using randomly amplified polymorphic DNA (RAPD) markers. Euphytica 102: 247–253.
-
- Degani C., Rowland L.J., Saunders J.A., Hokanson S.C., Ogden E.L., Golan-Goldhirsh A. and Galletta G.J. (2001) A comparison of genetic relationship measures in strawberry (Fragaria × ananassa Duch.) based on AFLPs, RAPDs, and pedigree data. Euphytica 117: 1–12.

