Integrating Gut Bacterial Diversity and Captive Husbandry to Optimize Vulture Conservation
- PMID: 32523573
- PMCID: PMC7261900
- DOI: 10.3389/fmicb.2020.01025
Integrating Gut Bacterial Diversity and Captive Husbandry to Optimize Vulture Conservation
Abstract
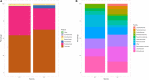
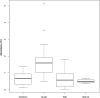
Endangered species recovery plans often include captive breeding and reintroduction, but success remains rare. Critical for effective recovery is an assessment of captivity-induced changes in adaptive traits of reintroduction candidates. The gut microbiota is one such trait and is particularly important for scavengers exposed to carcass microbiomes. We investigated husbandry-associated differences in the gut microbiota of two Old World vulture species using 16S RNA gene amplicon sequencing. Increased abundance of Actinobacteria occurred when vultures were fed quail but not rat or chicken. Conversely, diet preparation (sanitization) had no effect, although bacterial diversity differed significantly between vulture species, likely reflective of evolved feeding ecologies. Whilst the relative lack of influence of a sanitized diet is encouraging, changes in bacterial abundance associated with the type of prey occurred, representing a dietary influence on host-microbiome condition warranting consideration in ex situ species recovery plans. Incorporation of microbiome research in endangered species management, therefore, provides an opportunity to refine conservation practice.
Keywords: ex situ conservation; feeding ecology; gut microbiome; husbandry; old world vultures; prey diet; species recovery.
Copyright © 2020 Becker, Harrison, Whitehouse-Tedd, Budd and Whitehouse-Tedd.
Figures


Similar articles
-
Gut Microbiomes of Endangered Przewalski's Horse Populations in Short- and Long-Term Captivity: Implication for Species Reintroduction Based on the Soft-Release Strategy.Front Microbiol. 2020 Mar 12;11:363. doi: 10.3389/fmicb.2020.00363. eCollection 2020. Front Microbiol. 2020. PMID: 32226419 Free PMC article.
-
Comparative Study of Gut Microbiota in Wild and Captive Giant Pandas (Ailuropoda melanoleuca).Genes (Basel). 2019 Oct 20;10(10):827. doi: 10.3390/genes10100827. Genes (Basel). 2019. PMID: 31635158 Free PMC article.
-
Captivity and Animal Microbiomes: Potential Roles of Microbiota for Influencing Animal Conservation.Microb Ecol. 2023 Apr;85(3):820-838. doi: 10.1007/s00248-022-01991-0. Epub 2022 Mar 22. Microb Ecol. 2023. PMID: 35316343 Review.
-
Why Did the Bee Eat the Chicken? Symbiont Gain, Loss, and Retention in the Vulture Bee Microbiome.mBio. 2021 Dec 21;12(6):e0231721. doi: 10.1128/mBio.02317-21. Epub 2021 Nov 23. mBio. 2021. PMID: 34809450 Free PMC article.
-
Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates.Sci Total Environ. 2022 Sep 10;838(Pt 2):156178. doi: 10.1016/j.scitotenv.2022.156178. Epub 2022 May 23. Sci Total Environ. 2022. PMID: 35618126 Review.
Cited by
-
Captivity reduces diversity and shifts composition of the Brown Kiwi microbiome.Anim Microbiome. 2021 Jul 8;3(1):48. doi: 10.1186/s42523-021-00109-0. Anim Microbiome. 2021. PMID: 34238378 Free PMC article.
-
First metagenomic analysis of the Andean condor (Vultur gryphus) gut microbiome reveals microbial diversity and wide resistome.PeerJ. 2023 Jul 7;11:e15235. doi: 10.7717/peerj.15235. eCollection 2023. PeerJ. 2023. PMID: 37434868 Free PMC article.
-
Systematic mapping on the importance of vultures in the Indian public health discourse.Environ Sustain (Singap). 2022;5(2):135-143. doi: 10.1007/s42398-022-00224-x. Epub 2022 Apr 12. Environ Sustain (Singap). 2022. PMID: 37521585 Free PMC article. Review.
-
Metagenomic comparison of gut communities between wild and captive Himalayan griffons.Front Vet Sci. 2024 May 9;11:1403932. doi: 10.3389/fvets.2024.1403932. eCollection 2024. Front Vet Sci. 2024. PMID: 38784654 Free PMC article.
-
Gut microbiota of the threatened takahē: biogeographic patterns and conservation implications.Anim Microbiome. 2022 Jan 25;4(1):11. doi: 10.1186/s42523-021-00158-5. Anim Microbiome. 2022. PMID: 35078539 Free PMC article.
References
-
- Andrews S. (2010). FastQC: A Quality-Control Tool for High-Throughput Sequence Data. Cambridge, United Kingdom: Babraham Institute.
-
- Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75 129–137.
-
- Bangert R. L., Ward A. C. S., Stauber E. H., Cho B. R., Widders P. R. (1988). A survey of the aerobic bacteria in the feces of captive raptors. Avian Dis. 32 53–62. - PubMed
LinkOut - more resources
Full Text Sources

