Prevalence and Clonal Distribution of Azole-Resistant Candida parapsilosis Isolates Causing Bloodstream Infections in a Large Italian Hospital
- PMID: 32523896
- PMCID: PMC7261875
- DOI: 10.3389/fcimb.2020.00232
Prevalence and Clonal Distribution of Azole-Resistant Candida parapsilosis Isolates Causing Bloodstream Infections in a Large Italian Hospital
Abstract
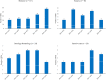
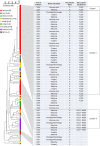
The most prevalent cause of nosocomial bloodstream infection (BSI) among non-C. albicans Candida species, Candida parapsilosis, may not only be resistant to azole antifungal agents but also disseminate to vulnerable patients. In this survey of BSIs occurring at a large Italian hospital between May 2014 and May 2019, C. parapsilosis accounted for 28.5% (241/844) of all Candida isolates causing BSI episodes. The majority of episodes (151/844) occurred in medical wards. Across the 5 yearly periods, the rates of azole non-susceptibility were 11.8% (4/34), 17.8% (8/45), 28.6% (12/42), 32.8% (19/58), and 17.7% (11/62), respectively, using the Sensititre YeastOne® method. Among azole non-susceptible isolates (54/241; 22.4%), 49 were available for further investigation. Using the CLSI reference method, all 49 isolates were resistant to fluconazole and, except one (susceptible dose-dependent), to voriconazole. Forty (81.6%) isolates harbored the Erg11p Y132F substitution and nine (18.4%) isolates the Y132F in combination with the Erg11p R398I substitution. According to their genotypes, as defined using a microsatellite analysis based on six short tandem repeat markers, 87.7% of isolates (43/49) grouped in two major clusters (II and III), whereas 4.1% of isolates (2/49) belonged to a separate cluster (I). Interestingly, all the isolates from cluster II harbored the Y132F substitution, and those from cluster III harbored both Y132F and R398I substitutions. Of 56 non-Italian isolates included as controls, two Indian isolates with the Y132F substitution had a genotype clearly differing from that of the isolates from clusters II and I. In conclusion, these findings show the dominance of clonal Y132F isolates in our hospital and suggest detection of the Y132F substitution as helpful tool to prevent transmission among hospitalized patients at risk of BSI.
Keywords: Candida parapsilosis; antifungal susceptibility; azole resistance; bloodstream infections; microsatellite genotyping; molecular clones.
Copyright © 2020 Martini, Torelli, de Groot, De Carolis, Morandotti, De Angelis, Posteraro, Meis and Sanguinetti.
Figures

References
-
- Asadzadeh M., Ahmad S., Al-Sweih N., Hagen F., Meis J. F., Khan Z. (2019). High-resolution fingerprinting of Candida parapsilosis isolates suggests persistence and transmission of infections among neonatal intensive care unit patients in Kuwait. Sci. Rep. 9:1340. 10.1038/s41598-018-37855-2 - DOI - PMC - PubMed
-
- Berkow E. L., Manigaba K., Parker J. E., Barker K. S., Kelly S. L., Rogers P. D. (2015). Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob. Agents Chemother. 59, 5942–5950. 10.1128/AAC.01358-15 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

